Figures & data
Table 1 The Summary of Studies Using CRISPR Tools to Target HCC
Table 2 CRISPR-Mediated Adoptive T Cell Immunotherapy Studies in HCC
Table 3 Clinical Trials Using CRISPR in Adoptive Cell Therapy for HCC
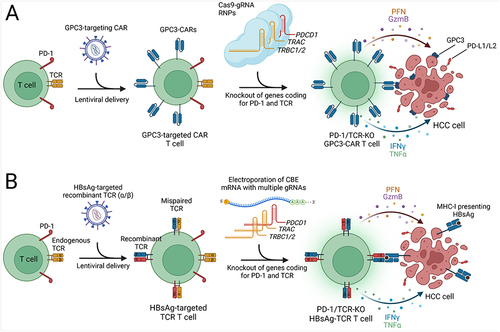
Figure 1 Overview of CRISPR-based gene editing technologies. Created with BioRender.com. (A) CRISPR-mediated approaches for gene disruption (NHEJ) and precise editing (HDR). (B) Double nickase strategy employing two Cas9 nickases for controlled and precise DSBs, minimizing off-target effects. (C) CRISPRa and CRISPRi systems for transient regulation of gene expression using deactivated Cas9 and transcription activator/inhibitor domains. (D) CRISPRoff system for durable gene repression by combining deactivated Cas9 with DNA methyltransferase domains and a transcriptional repressor domain. (E) Cytosine and adenine base editing systems that alter DNA bases without inducing double-strand breaks. (F) Prime editing method enabling precise installation of single base mutations and small indels using a Cas9 nickase and reverse transcriptase. (G) RNA-targeting Cas13 system for the manipulation of RNA molecules in cells.
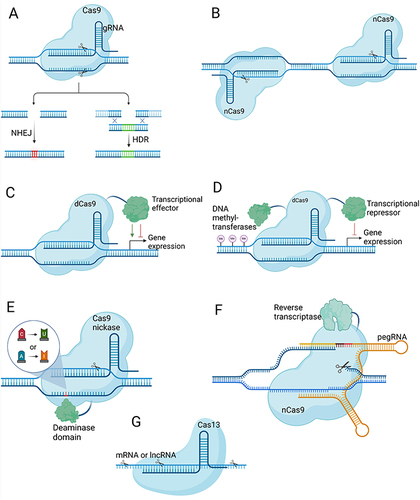
Figure 2 CRISPR-based enhancements for CAR T and TCR T cell therapies in HCC. Created with Biorender.com. (A) Autologous T cells are transduced with a lentivirus carrying a GPC3-targeting CAR expression cassette, followed by electroporation of Cas9-gRNA RNPs to knockout genes encoding the PD-1 and TCR α and β chains. These modifications prevent T cell exhaustion and enhance specificity. The engineered CAR T cells effectively bind GPC3 on HCC cells via GPC3-targeted CARs, triggering a release of cytotoxic proteins (perforin, granzyme B) and pro-inflammatory cytokines (IFN-γ, TNF-α), which collectively contribute to the targeted destruction of tumor cells. (B) Development of Hepatitis B surface antigen (HBsAg)-targeted TCR T cells involves lentiviral delivery of recombinant TCR α and β chains to T cells. The presence of endogenous TCR chains can lead to the formation of mispaired TCRs with unpredictable antigen specificity, posing a potential safety risk. For enhancing safety and efficacy, the cytosine base editor (CBE) is introduced as mRNA to perform knockout of endogenous TCR genes and PD-1. The specific interaction between the HBsAg-targeted TCR and HBsAg presented by MHC-I on HCC cells activates the TCR T cells, promoting robust antitumor activity.