Figures & data
Table 1 Methylation-Related Biomarkers in HCC
Table 2 Effect of DNA Promoter Methylation Modification of Diverse Genes on HCC
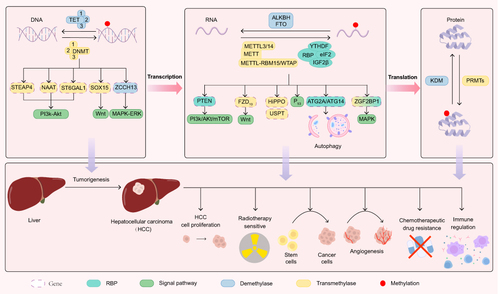
Figure 1 HCC methylation occurs in DNA, RNA, and protein. M6A modifications are regulated by a family of methyltransferases (writer), demethylases (eraser), and specific RNA binding proteins(reader). The methyltransferase family includes METTL family and WTAP. Demethylases include FTO, ALKBH5 meanwhile others, which directly remove m6A modifications from mRNAs. Specific RNA binding proteins consist of YTH domain proteins, the IGF binding protein 2 family, eukaryotic initiation factor 3, and nuclear heterogeneous protein2β1.