Figures & data
Figure 1 TALEN and CRISPR targets in NF-κB genes and HDR donor. (A) The selected left and right homologous (hom) arms and TALEN and CRISPR targets in the NF-κB family genes. Up strand: sense strand (from left to right: 5′ to 3′); down strand: anti-sense strand (from left to right: 3′ to 5′); bases labeled with a solid arrow: left hom arm/primer F and right hom arm; bases in blue; TALEN target sites: bases labeled with dotted arrow: sgRNA target sites; bases in red: stop codon; bases in square: PAM. (B) The HDR donor containing Gslink and ZsGreen coding sequences and ending with homology arms targeting to five NF-κB family genes. The length of left and right hom arms is 35 nt.
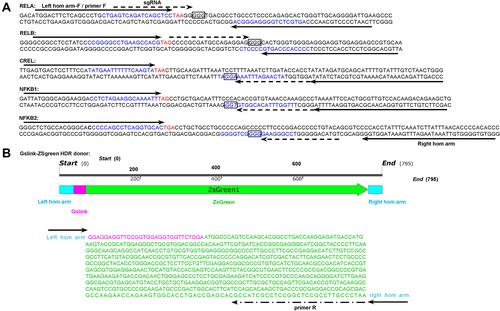
Figure 2 Editing NF-κB genes in three cell lines with TALEN and CRISPR. Linear dsDNA fragments containing a Gslink-ZsGreen-coding sequence and ending with homology arms homologous to NF-κB family genes were used as HDR donors. (A) Representative fluorescent images of 293T cells after the TALEN and CRISPR editing and HDR. The representative fluorescent images of HepG2 and PANC1 cells after the TALEN and CRISPR editing and HDR are shown in Figure S1A and S2A. The scale bar is 100 μm. (B) Flow cytometry assay of 293T cells after the TALEN and CRISPR editing and HDR. The flow cytometry assays of HepG2 and PANC1 cells after the TALEN and CRISPR editing and HDR are shown in Figure S1B and S2B. (C) The statistical analysis of flow cytometry assays of three biological replicates. The results are shown as mean values ± standard deviation (SD). The Student t test was performed to assess the statistical significance. ***p<0.001,**p<0.01.
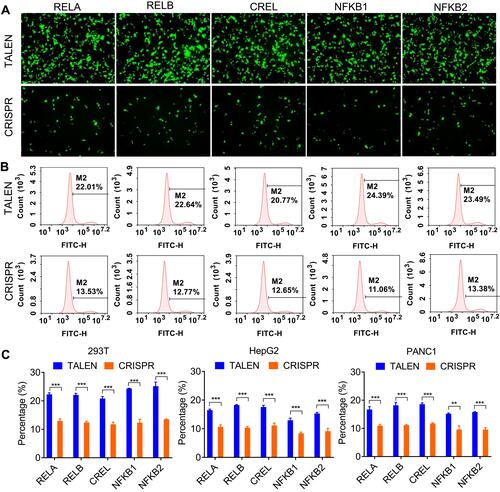
Figure 3 Detection of editing efficiency of TALEN and CRISPR with qPCR. A same amount of gDNA (100 ng) from all cells after the TALEN and CRISPR editing and HDR was used to detect the fused NF-κB-ZsGreen genes by qPCR. (A) The amplification plots of fused NF-κB-ZsGreen genes. (B) Visualization of qPCR products with agarose gel electrophoresis. (C) The statistical analysis of qPCR detection. ΔCt was calculated by CtNF-κB-ZsGreen-CtGAPDH. The results are shown as mean values ± standard deviation (SD). The Student t test was performed to assess the statistical significance. ***p<0.001.
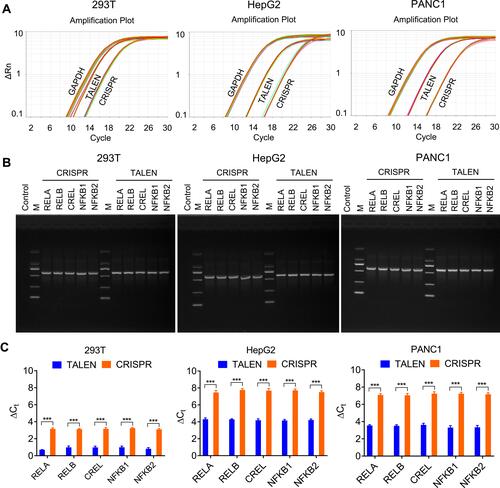
Figure 4 TNFα stimulation to the TALEN-edited cells. The TALEN-edited cells were treated by TNFα, an NF-κB stimulator, and the nuclei were isolated from the treated cells. (A) Representative fluorescent images of the isolated nuclei of 293T cells. The representative fluorescent images of the isolated nuclei of HepG2 and PANC1 cells are shown in Figure S3A and S4A. The scale bar is 100 μm. (B) Flow cytometry assay of the isolated nuclei of 293T cells. The flow cytometry assays of the isolated nuclei of HepG2 and PANC1 cells are shown in Figure S3B and S4B.
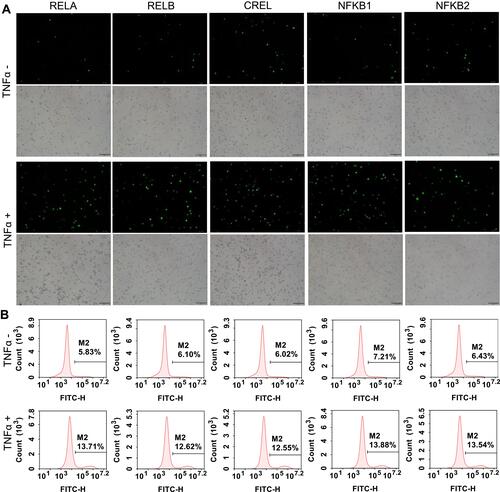
Figure 5 Fluorescent imaging of flow cytometry-sorted positive cells. (A) 293T cells. (B) HepG2 cells. (C) PANC1cells. The scale bar is 100 μm. The flow cytometry assays of the sorted cells are shown in Figure S5.
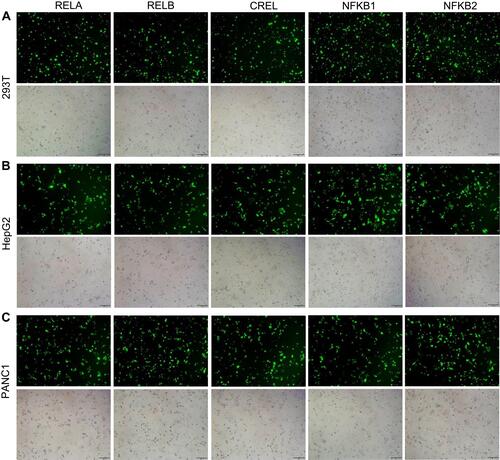
Figure 6 BAY 11–7082 treatment to the flow cytometry-sorted positive cells. (A) Representative fluorescent images of BAY 11–7082-treated and -untreated 293T cells. The representative fluorescent images of the BAY 11–7082-treated and -untreated HepG2 and PANC1 cells are shown in Figure S6A and S7A. The scale bar is 100 μm. (B) Flow cytometry assay of BAY 11–7082-treated and –untreated 293T cells. The flow cytometry assay of BAY 11–7082-treated and -untreated HepG2 and PANC1 cells are shown in Figure S6B and S7B.
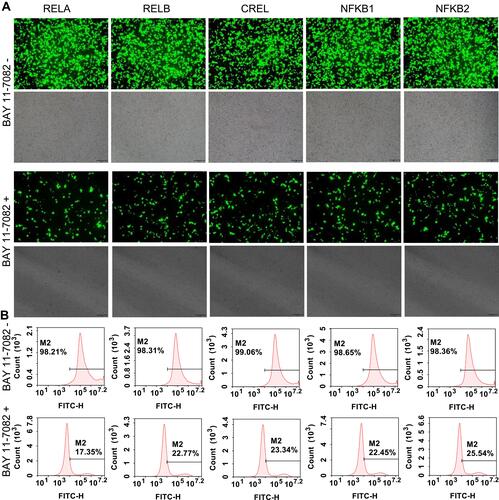
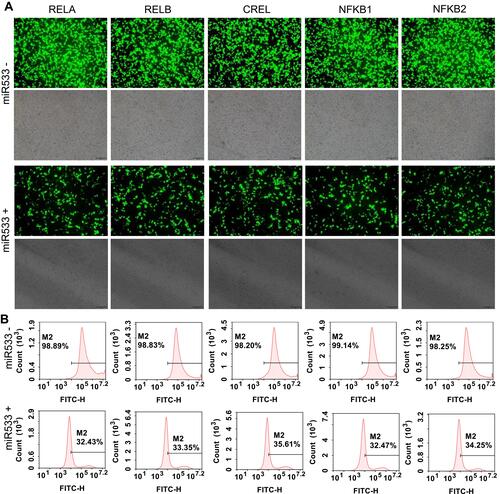
Figure 7 MiR533 treatment to the flow cytometry-sorted positive cells. (A) Representative fluorescent images of miR533-treated and -untreated 293T cells. The representative fluorescent images of the miR533-treated and -untreated HepG2 and PANC1 cells are shown in Figure S8A and S9A. The scale bar is 100 μm. (B) Flow cytometry assay of miR533-treated and –untreated 293T cells. The flow cytometry assay of miR533-treated and -untreated HepG2 and PANC1 cells are shown in Figure S8B and S9B.