Figures & data
Figure 1 Effect of a high fat diet of pregnant rats on kidney growth of offspring. (A) Body weight, kidney weight, kidney index, and systolic blood pressure (SBP) at the indicated time points in both groups. (B) Representative PAS-stained kidney sections and a graph of nephron counts. Images were captured at a magnification of 200×; scale bar = 100 μm. (n =6 per group, *p < 0.05 vs control). (C) Western blot analysis of AQP1 proteins (n =7 per group, *p < 0.05 vs control). (D) Immunofluorescence (IF) staining of vimentin in both groups at E20 or 12W (magnification of 800×; scale bar = 50 μm).
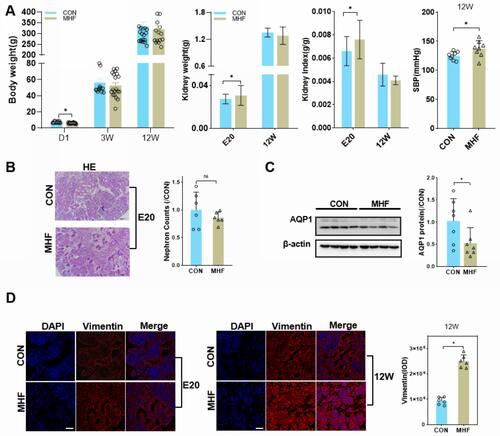
Figure 2 Maternal high fat (MHF) diet induced dysregulation of peroxins (PEXs) in the fetal kidney. (A) Western blot analysis of PEXs proteins. (B) qPCR analysis of the PEXs mRNA levels. (n =7-8 per group, *p < 0.05 vs control). (C) IF staining of PEX14 and catalase colocalization and distribution of PEX3 in the fetal kidney (magnification of 800×; scale bar = 50 μm).
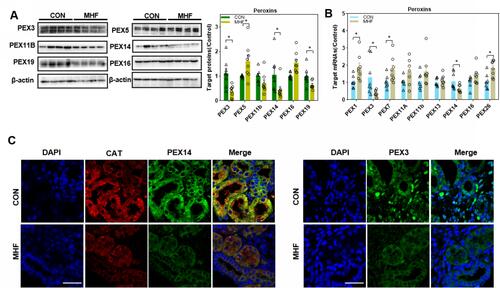
Figure 3 Effects of maternal high-fat on fatty acid β-oxidation (FAO) regulators in the fetal kidney. (A) Western blot analysis of FAO regulators. (B) The qPCR analysis of the mRNA levels of FAO regulators. (n =8 per group, *p < 0.05 vs control). (C) IF staining of Peroxisomal membrane protein (PMP) 70 and catalase (CAT) co-localization in the fetal kidney (magnification of 800×; scale bar = 50μm).
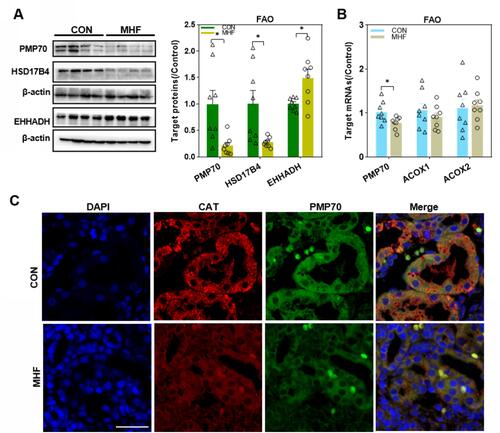
Figure 4 Effects of MHF on antioxidants in the fetal kidney. (A) The qPCR analysis of the mRNA level of antioxidants in both groups. (B) Protein expression of the antioxidants CAT and superoxide dismutase (SOD)2. (n = 7–8 per group, *p < 0.05 vs control). (C) IF staining showing the locations and expression of nitric oxide synthase (NOS)2 in the fetal kidney of CON and MHF group (400×, scale bar = 50μm, n=6 per group, *p < 0.05 vs control). (D) MDA levels in the fetal kidney of both groups. (n = 5 per group, *p < 0.05 vs control).
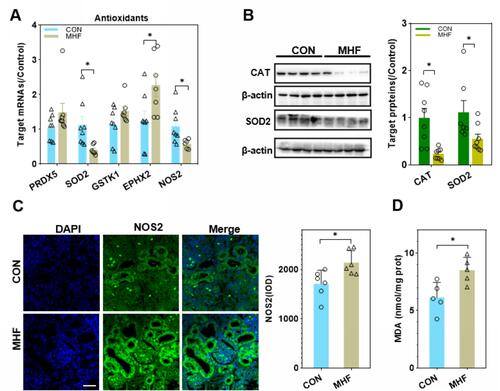
Figure 5 Protein interaction analysis. (A) STRING Protein–Protein Interaction network involving the detected differentially expressed genes. (B) KEGG pathway analysis of differentially expressed genes enriched in peroxisome-related pathways. Red indicates an increase in expression and green, a decrease.
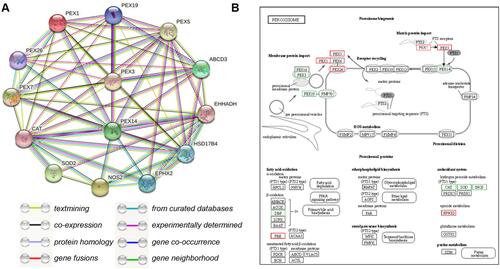
Figure 6 MHF triggered NRF2 activation in the fetal kidney. (A) Western blot analysis of the expression of Kelch-like ECH associated protein (KEAP)1 and nuclear translocation of NF-E2- related factor 2 (NRF2). (B) qPCR analysis of the mRNA levels of NRF2 and associated downstream genes. *p < 0.05 vs control; (n = 7–8 per group, *p < 0.05 vs control).
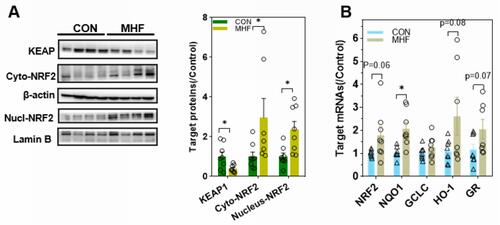
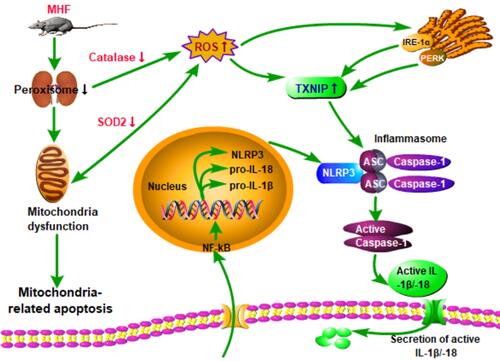
Figure 7 MHF induced activation of the NLR family, pyrin domain-containing (NLRP)3 inflammasome in the fetal kidney. (A) The qPCR analysis of mRNA levels of genes associated with the inflammasome in the fetal kidney. (B) Expression levels of protein associated with the NLRP3 inflammasome in the fetal kidney. (n = 7–8 per group, *p < 0.05 vs control). (C) IHC of NLRP3 and CD68 and IF staining of CD68 showed the expression and location of NLRP3, and CD68 in the fetal kidney (magnification of 400×; scale bar = 50μm).
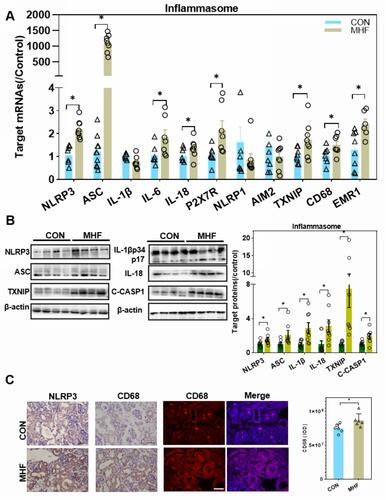
Figure 8 Effects of MHF on endoplasmic reticulum stress (ERS) and apoptosis regulators in the fetal kidney. (A) Protein expression levels of ERS regulators in the fetal kidney. (n = 7–8 per group, *p < 0.05 vs control). (B) The qPCR analysis of the mRNA levels of ERS regulators (C) IF staining showing the expression and location of XBP-1 (400×, scale bar = 50μm, n=6 per group, *p < 0.05 vs control). (D) The qPCR analysis of the mRNA levels of Bax and BCL-2 in the fetal kidney. (E) Protein expression of apoptosis regulators in the fetal kidney. (n = 7–8 per group, *p < 0.05 vs control).
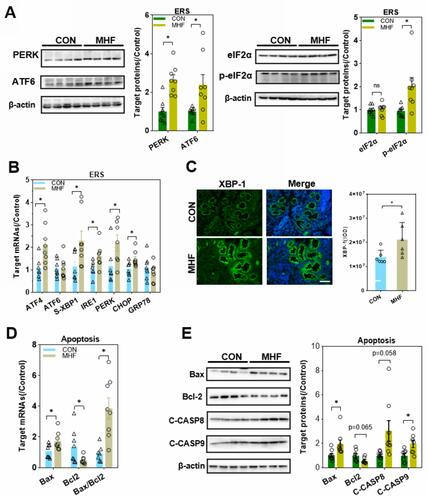
Figure 9 Effect of MHF on TECs in fetus. The qPCR analysis of the mRNA levels of PEXs/antioxidants/inflammasome in the fetal kidney. (n = 3–6 per group, *p < 0.05 vs control).
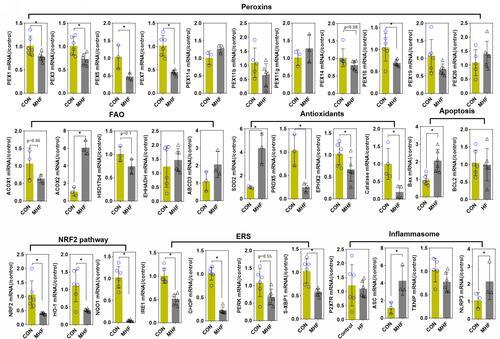
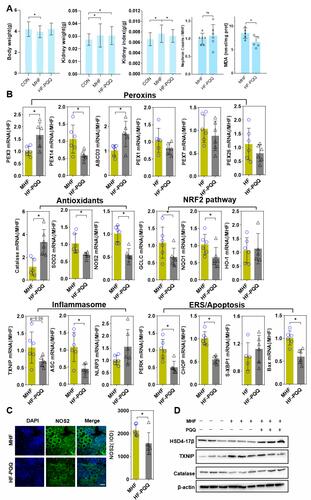
Figure 10 Effect of PQQ on renal programming. (A) Body weight, kidney weight, kidney index, and MDA level in the fetus after PQQ treatment. (B) The qPCR analysis of the mRNA level of PEXs, antioxidants, NRF2 pathway and inflammasome genes in the fetus after PQQ treatment. (n = 6 per group, *p < 0.05 vs MHF). (C) IF staining showing the expression of NOS2 in the fetal kidney of MHF and HF-PQQ group (400×, scale bar = 50μm, n=6 per group, *p < 0.05 vs MHF). (D) Western blot analysis of the expression of Hsd17b4, Txnip, and catalase in fetal kidney.