Figures & data
Table 1 Relationship Between Circ-PRKDC Expression and Clinicopathologic Features of Colorectal Cancer Patients
Figure 1 High expression of circ-PRKDC was observed in 5-FU-resistant CRC tissues and cells. (A) Circ-PRKDC expression in 5-FU-resistant and 5-FU-sensitive CRC tissues was determined by qRT-PCR. (B) Circ-PRKDC expression in FHC, SW620, SW620/5-FU, SW480 and SW480/5-FU cells was measured by qRT-PCR. (C and D) The expression of circ-PRKDC and linear PRKDC in SW480 and SW480/5-FU cells treated with or without RNase R were measured by qRT-PCR assay. *P<0.05.
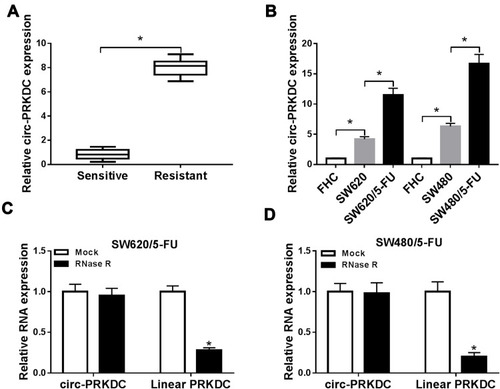
Figure 2 Downregulation of circ-PRKDC improved 5-FU sensitivity in 5-FU-resistant CRC cells. (A) IC50 of 5-FU in SW620, SW620/5-FU, SW480 and SW480/5-FU cells was examined by CCK-8 assay. (B–G) SW620/5-FU and SW480/5-FU cells were transfected with si-PRKDC or si-NC. (B) The expression of circ-PRKDC in SW620/5-FU and SW480/5-FU cells was determined by qRT-PCR. (C) IC50 of 5-FU in SW620/5-FU and SW480/5-FU cells was evaluated by CCK-8 assay. (D) The colony formation ability of SW620/5-FU and SW480/5-FU cells was analyzed by colony formation assay. (E) The invasion of SW620/5-FU and SW480/5-FU cells was assessed by transwell assay. (F and G) The protein levels of P-gp and MRP1 in SW620/5-FU and SW480/5-FU cells were measured using Western blot assay. *P<0.05.
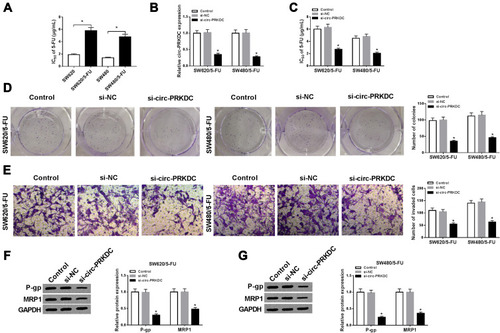
Figure 3 Circ-PRKDC targeted miR-375 to regulate 5-FU resistance in 5-FU-resistant CRC cells. (A) The binding sites between circ-PRKDC and miR-375 were predicted by starBase v2.0. (B and C) The combination between circ-PRKDC and miR-375 was investigated by dual-luciferase reporter assay. (D and E) After RIP assay, the expression levels of circ-PRKDC and miR-375 in IgG or Ago2 immunoprecipitates were measured by qRT-PCR analysis. (F) MiR-375 expression in 5-FU-resistant and 5-FU-sensitive CRC tissues was measured using qRT-PCR assay. (G) MiR-375 expression in FHC, SW620, SW480, SW620/5-FU and SW480/5-FU cells was determined by qRT-PCR. (H) The correlation between circ-PRKDC and miR-375 was analyzed by Spearman correlation coefficient analysis. (I and J) The expression levels of circ-PRKDC and miR-375 in SW620/5-FU and SW480/5-FU cells transfected with si-NC, si-circ-PRKDC, pcDNA or circ-PRKDC were examined by qRT-PCR. (K–P) SW620/5-FU and SW480/5-FU cells were assigned to control, si-NC, si-circ-PRKDC, si-circ-PRKDC+anti-miR-NC and si-circ-PRKDC+anti-miR-375 groups. (K) The expression of miR-375 in SW620/5-FU and SW480/5-FU cells was examined through qRT-PCR. (L) IC50 of 5-FU in SW620/5-FU and SW480/5-FU cells was evaluated by CCK-8 assay. (M) The colony numbers of SW620/5-FU and SW480/5-FU cells were tested by colony formation assay. (N) The invasion of SW620/5-FU and SW480/5-FU cells was detected via transwell assay. (O and P) The protein levels of P-gp and MRP1 in SW620/5-FU and SW480/5-FU cells were measured via Western blot assay. *P<0.05.
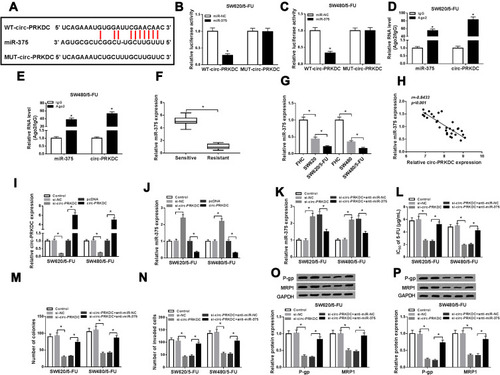
Figure 4 FOXM1 was a direct target gene of miR-375. (A) The potential binding sites between miR-375 and FOXM1 were presented. (B and C) The relationship between miR-375 and FOXM1 was verified by dual-luciferase reporter assay. (D and E) The interaction between FOXM1 and miR-375 were analyzed by RIP assay and qRT-PCR analysis. (F and G) The mRNA and protein levels of FOXM1 in 5-FU-resistant and 5-FU-sensitive CRC tissues were measured by qRT-PCR assay and Western blot assay, respectively. (H) The correlation between miR-375 and FOXM1 in CRC tissues was analyzed by Spearman correlation coefficient analysis. (I and J) The mRNA and protein levels of FOXM1 in FHC, SW620, SW480, SW620/5-FU and SW480/5-FU cells were measured by qRT-PCR assay and Western blot assay, respectively. *P<0.05.
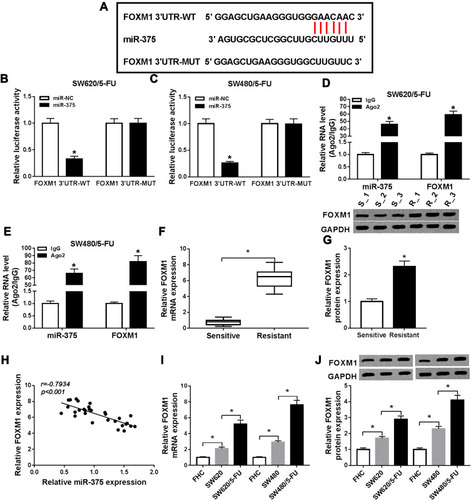
Figure 5 MiR-375 inhibited 5-FU resistance by binding to FOXM1 in 5-FU-resistant CRC cells. SW620/5-FU and SW480/5-FU cells were divided into 5 groups: control, miR-NC, miR-375, miR-375+pcDNA and miR-375+FOXM1. (A and B) The protein level of FOXM1 in SW620/5-FU and SW480/5-FU cells was measured by Western blot assay. (C) IC50 of 5-FU in SW620/5-FU and SW480/5-FU cells was evaluated by CCK-8 assay. (D) The colony formation ability of SW620/5-FU and SW480/5-FU cells was analyzed by colony formation assay. (E) SW620/5-FU and SW480/5-FU cell invasion was explored by transwell assay. (F and G) The protein levels of P-gp and MRP1 in SW620/5-FU and SW480/5-FU cells were detected by Western blot assay. *P<0.05.
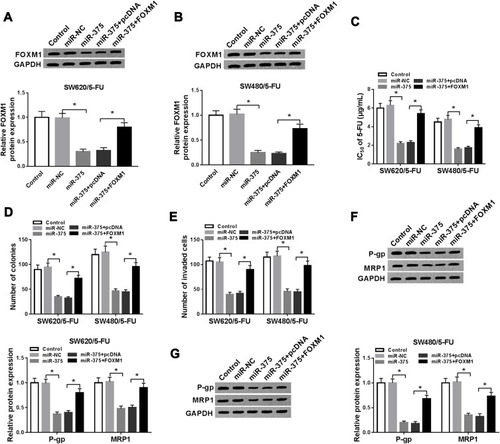
Figure 6 Silencing of circ-PRKDC downregulated FOXM1 expression via binding to miR-375 in 5-FU-resistant CRC cells. (A and B) SW620/5-FU and SW480/5-FU cells were transfected with si-NC, si-circ-PRKDC, si-circ-PRKDC+anti-miR-NC or si-circ-PRKDC+anti-miR-375 and then the protein level of FOXM1 was determined by Western blot assay. *P<0.05.
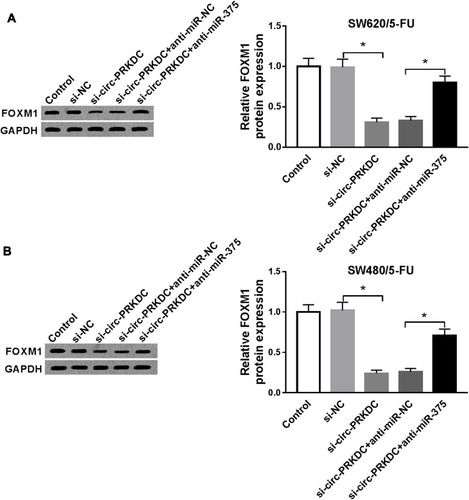
Figure 7 Circ-PRKDC knockdown suppressed wnt/β-catenin pathway in 5-FU-resistant CRC cells by regulating miR-375 and FOXM1. (A and B) SW620/5-FU and SW480/5-FU cells were assigned to control, si-NC, si-circ-PRKDC, si-circ-PRKDC+anti-miR-NC, si-PRKDC+anti-miR-375, si-circ-PRKDC+pcDNA and si-circ-PRKDC+FOXM1 groups and then the protein levels of β-catenin and c-Myc were examined by Western blot assay. *P<0.05.
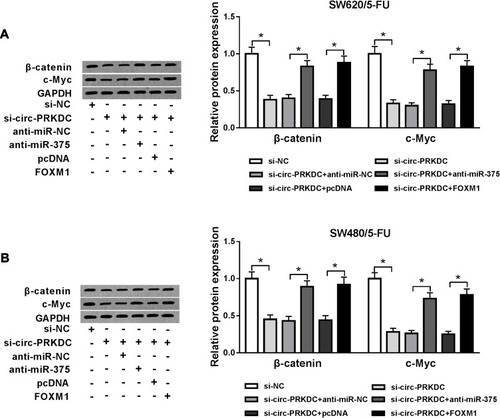
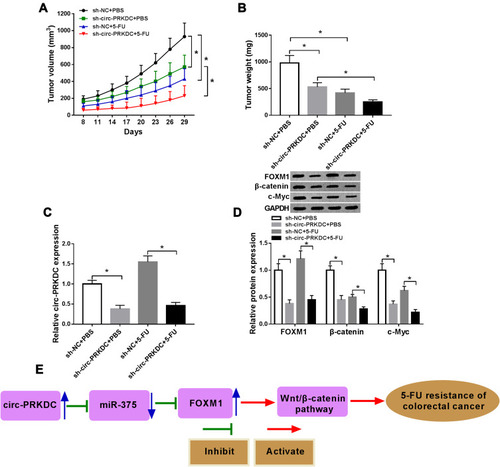
Figure 8 Knockdown of circ-PRKDC improved 5-FU sensitivity of CRC in vivo. Sh-NC or sh-circ-PRKDC transfected SW480/5-FU cells were injected into the mice and treated with PBS or 5-FU every 3 days after 8 days. (A) Tumor volume was examined every 3 days after 8 days. (B) Tumor weight was measured after 29 days. (C) The expression of circ-PRKDC in the harvested tissues was determined by qRT-PCR. (D) The protein levels of FOXM1, β-catenin and c-Myc in the harvested tissues were determined by Western blot assay. (E) The schematic representation of circ-PRKDC/miR-375/FOXM1/wnt/β-catenin pathway in the modulation of 5-FU resistance in CRC. *P<0.05.