Figures & data
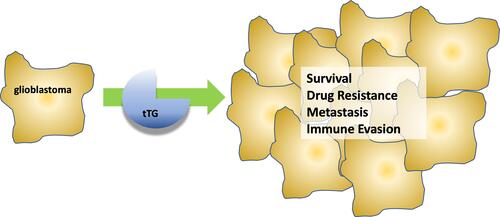
Figure 1 Structure of tissue transglutaminase. (A) Crystal structures of tTG in the closed-state conformation (left, PDB ID 1KV3), and the open-state conformation (right, PDB ID 2Q3Z). Boxes show the binding site for nucleotide or crosslinking substrate, respectively. Addition of calcium drives tTG to the open state, while addition of nucleotide stabilizes the closed state. (B) Zoomed in image of the nucleotide binding pocket of closed-state tTG. Shown in black and colored by heteroatom is the nucleotide GTP. The GTP bound state (PDB 4PYG) is colored in blue and the GDP bound state (1KV3) is colored in grey. GDP is omitted from the nucleotide binding site for clarity, since the position of the nucleotide does not change between states. Amino acid side chains that form hydrogen bonds with the nucleotide are labeled and drawn as sticks. (C) Zoomed in image of the peptide binding site of open-state tTG (PDB 2Q3Z). Shown in green is an irreversible gluten peptide mimetic, Ac-P(DON)LPF-NH2. The highly conserved catalytic triad (C277, H335, D358) is labeled and shown as sticks, with the addition of W241 which is also conserved among transglutaminases.
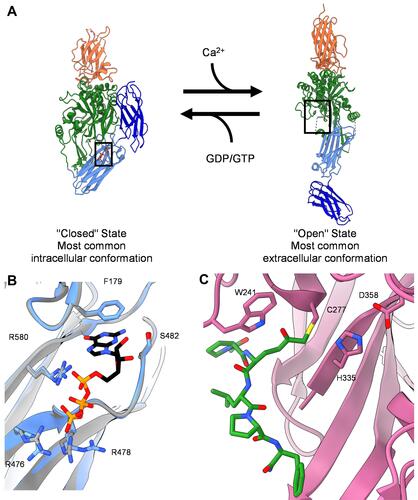
Figure 2 Consensus sequence for tTG crosslinking substrates. Alignment of all glutamine-donor crosslinking substrates described in the Transdab database demonstrates that other than glutamine (Q at position 0), there is little to no favoritism for different residues at up to five positions before or after the glutamine residue to be crosslinked. Figure generated with Seq2Logo 2.0.
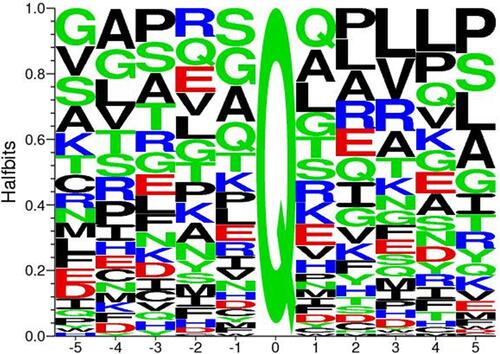
Figure 3 Mechanisms by which tissue transglutaminase impacts glioblastoma aggressiveness. (A) Without tTG, in many glioblastoma cells c-Cbl ubiquitinates the EGFR, priming it for degradation. When tTG is present, it sequesters the c-Cbl, preventing ubiquitination and effectively stabilizing the growth receptor, functionally increasing its expression. (B) Extracellular tTG is able to crosslink fibronectin displayed on the outside of microvesicles (MVs) to integrins expressed on cell surfaces. This allows the MVs to dock to the cells, and release their contents.
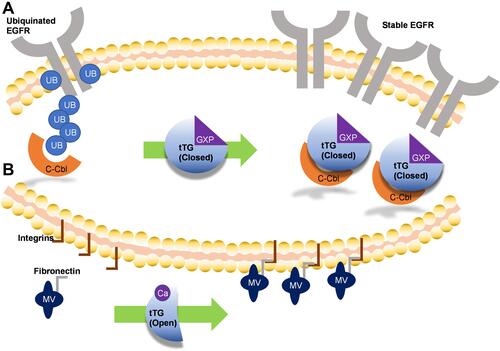
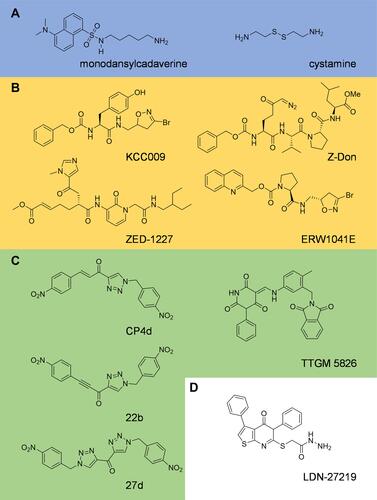
Figure 4 Inhibitors of tissue transglutaminase. Tissue transglutaminase inhibitors can be broadly classified as (A) alternate substrates, an older class of inhibitor which takes the place of a natural substrate of tTG, thus blocking biologically relevant crosslinking reactions. (B) Irreversible substrate competitive inhibitors, which bind to the crosslinking substrate site and covalently attach to tTG, blocking crosslinking. These tend to be derived from peptide structures. (C) Reversible substrate competitive inhibitors. These molecules bind to the substrate crosslinking site of tTG, but do not modify the enzyme. (D) Inhibitors which bind to the nucleotide binding pocket. Relatively few inhibitors utilize this strategy, but they stabilize the closed-state of tTG, thus blocking crosslinking activity.