Figures & data
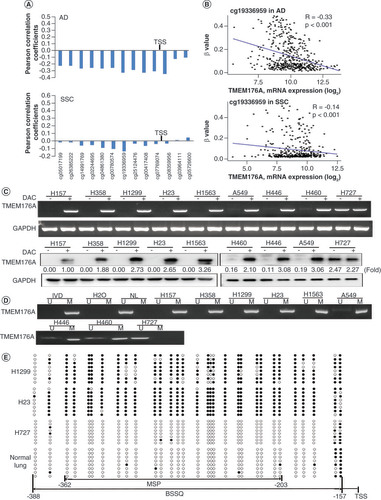
(A) The association of TMEM176A expression and methylation at each CpG site of promoter region. (B) Scatter plots show reduced TMEM176A expression is associated with methylation status of cg19336959 in 457 cases of lung adenocarcinoma and 372 cases of lung squamous cell carcinoma. (C) Semiquantitative reverse transciptase PCR and western blot show the levels of TMEM176A expression in lung cancer cells (H157, H358, H1299, H23, H1563, A549, H446, H460 and H727) both in RNA and protein levels. (D) Methylation status of TMEM176A in lung cancer cells. (E) BSSQ results in H1299, H23, H727 cells and normal lung. Filled circles: methylation; open circles: unmethylation.
***p < 0.001.
BSSQ: Sodium bisulfite sequence; (-): without DAC; (+): with DAC; AD: Lung adenocarcinoma; GAPDH: Internal control; H2O: Double-distilled water; IVD: Invitral methylated DNA; M: Methylation; MSP: Methylation-specific PCR; NL: Normal lymph cell DNA; SCC: Lung squamous cell carcinoma; TSS: Transcription start site; U: Unmethylation.
(A) Representative methylation-specific PCR results. (B) TMEM176A staining in lung cancer and adjacent tissue samples (top: 200×; bottom: 400×). (C) Box plots for TMEM176A expression, horizontal lines represent the median score; the bottom and top of the boxes represent the 25th and 75th percentiles, respectively; vertical bars represent expression levels. (D) Bar diagram: the levels of TMEM176A expression and methylation status.
*p < 0.05; ***p < 0.001.
IVD: In vitro methylated DNA; LC: Primary lung cancer; M: Methylation; N: Normal lung tissue samples; NL: Normal lymph cell DNA; U: Unmethylation.
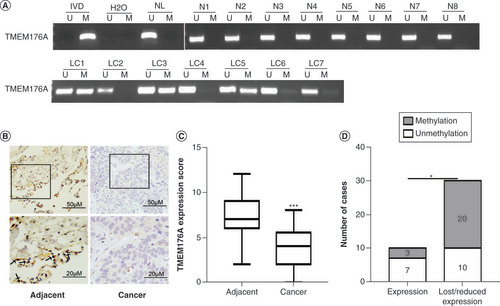
Table 1. The association of TMEM176A methylation and clinical factors in non-small-cell lung cancer.
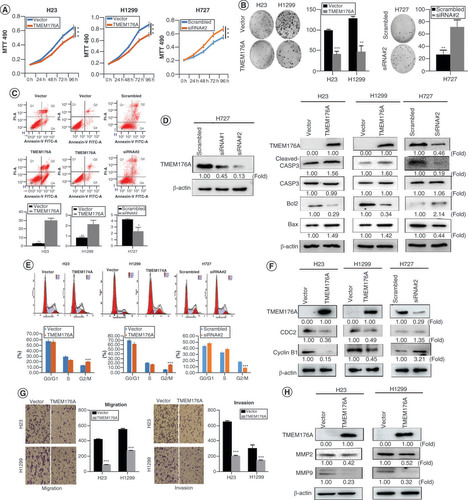
(A) Growth curves represent cell viability. (B) Colony formation results. Bar diagram: average number of tumor clones. (C) Flow cytometry shows apoptosis results. (D) The effect of TMEM176A on caspase-3, cleaved caspase-3, Bcl2 and Bax expression in H23, H1299 and H727 cells. siRNA#1 and siRNA#2: siRNA for TMEM176A. (E) Cell phase distribution in TMEM176A unexpressed and expressed H23, H1299 and H727 cells. (F) The effect of TMEM176A on CDC2 and cyclin B1 expression in H23, H1299 and H727 cells. (G) The migration and invasion assays for TMEM176A expressed and unexpressed H23 and H1299 cells. (H) The effect of TMEM176A on MMP-2 and MMP-9 expression in H23 and H1299 cells.
*p < 0.05; **p < 0.01; ***p < 0.001.
β-actin: Internal control; Scrambled: Scrambled siRNA serve as negative control; siRNA#2: siRNA for TMEM176A; TMEM176A: TMEM176A expressing vector; Vector: Control vector.
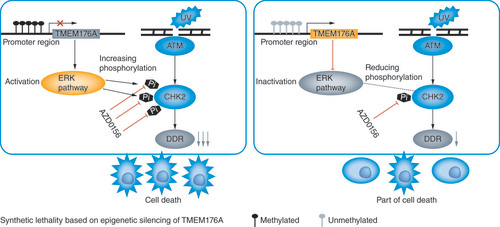
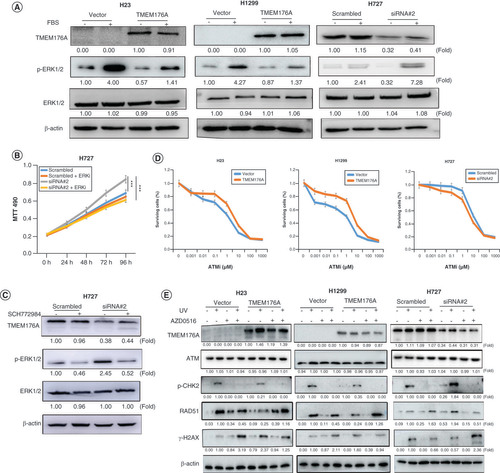
(A) western blots results. β-actin: internal control. (-): no serum stimulation; (+): serum stimulation. (B) Growth curves represent cell viability. SiRNA#2: siRNA for TMEM176A. (C) western blot results. SCH772984: p-ERK1/2 inhibitor; (-): absence of SCH772984; (+): presence of SCH772984. (D) MTT assays show OD value for IC50. ATMi: ATM inhibitor. (E) Epigenetic silencing TMEM176A sensitized lung cancer cells to UV and AZD0156. western blot shows the levels of TMEM176A, ATM, p-CHK2, γ-H2AX, RAD51 and β-actin.
***p < 0.001.
AZD0156: ATM inhibitor; (-): absence of AZD0156 or UV; (+): presence of AZD0156 or UV.
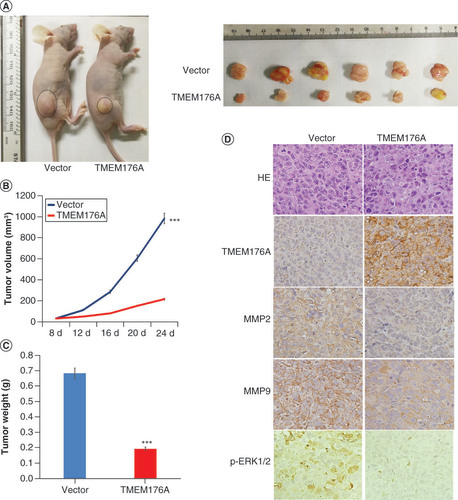
(A) TMEM176A unexpressed and re-expressed H1299 cell xenografts in mice. (B) Growth curves for TMEM176A unexpressed and re-expressed H1299 cell xenografts. (C) Tumor weights: TMEM176A unexpressed and re-expressed H1299 cell xenografts. (D) TMEM176A affects the levels of MMP-2, MMP-9 and p-ERK1/2 in H1299 cell xenografts.
***p < 0.001.