Figures & data
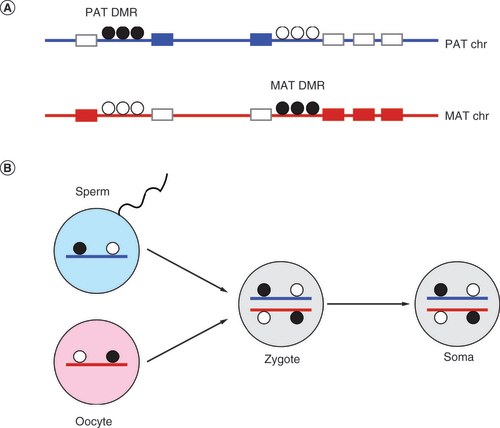
(A) Imprinted domains. Imprinted genes (rectangles) are expressed either in the paternally (blue) or in the maternally (red) inherited chromosome. Two imprinted domains are shown in one chromosome; one domain has a paternal and the other has a maternal germline DMR (gDMR). Dots indicate a group of methylated (black) or unmethylated (white) CpG dinucleotides in the DMRs. Silent gene alleles are shown by grey rectangles. (B) The imprinted gDMRs, which control imprinted gene expression in the soma, originate in the parental germlines. One paternal and one maternal gDMR is depicted as they arrive into the zygote methylated from the sperm or the egg, respectively.
DMR: Differentially methylated regioN; MAT: Maternal; PAT: Paternal.
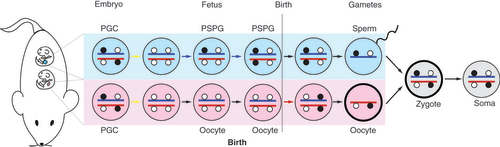
PGC undergo erasure of imprints in both the male and female germlines in the embryo before they attain the new gamete-specific imprints. The new paternal germline differentially methylated regions are set up in both chromosomes of PSPG in the male fetus. Maternal germline differentially methylated regions are set up in both chromosomes of growing oocytes in the female after birth. The male and female germline events are shown in two fetuses, one male and one female, for simplicity.
PGC: Primordial germ cells; PSPG: Prospermatogonia.
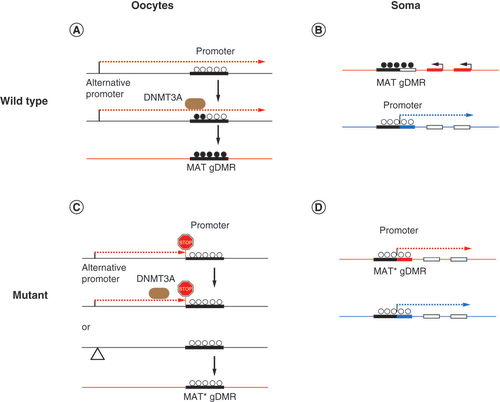
(A) Transcription across the MAT gDMR occurs from an oocyte-specific alternative promoter, facilitating de novo DNA methylation of the MAT gDMR by DNMT3A. (B) In the soma, a typical MAT gDMR is methylated in the chromosome inherited from the oocyte (top). The same sequence is unmethylated and functions as a promoter in the paternal chromosome only and affects other imprinted genes in the domain (bottom). (C) Truncation of the alternative transcript or deletion of the oocyte-specific promoter results in hypomethylated MAT gDMR (MAT*). (D) When the MAT* gDMR is inherited from the female germline, imprinted gene expression in that domain is absent in the soma.
gDMR: Germline differentially methylated regions; MAT: Maternal.
Schematic summary of the relationship between gDMRs and transcription in prospermatogonia. In male germ cells, broad, low-level RNA runs across a paternal gDMR, which becomes de novo methylated between 15 and 17 days of gestation. Such transcripts (dashed arrows) are globally found in male germ cells at 15.5 dpc but not in male somatic cells of the gonad, which only display regular transcripts (solid arrows). Transcription initiates from a maternal gDMR, which displays H3K4me2/3 enrichment and is likely protected from de novo methylation by H3K4 methylation in prospermatogonia. At the same time the MAT gDMR lacks H3K36me3 in prospermatogonia [Citation23].
gDMR: germline differentially methylated regions; MAT: Maternal; PAT: Paternal.
![Figure 4. Broad, low-level RNA crosses paternal germline differentially methylated regions in mouse prospermatogonia at the time of de novo DNA methylation.Schematic summary of the relationship between gDMRs and transcription in prospermatogonia. In male germ cells, broad, low-level RNA runs across a paternal gDMR, which becomes de novo methylated between 15 and 17 days of gestation. Such transcripts (dashed arrows) are globally found in male germ cells at 15.5 dpc but not in male somatic cells of the gonad, which only display regular transcripts (solid arrows). Transcription initiates from a maternal gDMR, which displays H3K4me2/3 enrichment and is likely protected from de novo methylation by H3K4 methylation in prospermatogonia. At the same time the MAT gDMR lacks H3K36me3 in prospermatogonia [Citation23].gDMR: germline differentially methylated regions; MAT: Maternal; PAT: Paternal.](/cms/asset/27c3f99e-7c5e-4968-b937-fc6559606572/iepi_a_12366747_f0004.jpg)
(A) Broad, low-level transcription runs across a PAT gDMR and facilitates de novo DNA methylation of the PAT gDMR [Citation1] by DNMT3A. (B) A typical PAT gDMR is methylated in the chromosome inherited from the sperm (bottom). The same sequence is unmethylated and functions (in this case, as an enhancer-insulator bound by CTCF, yellow) exclusively in the maternal chromosome (top), affecting imprinted genes of the domain. (C) Truncation of the broad, low-level transcript results in hypomethylated PAT* gDMR. (D) When a hypomethylated PAT* gDMR is inherited from the male germline, imprinted gene expression is absent in the soma.
gDMR: Germline differentially methylated regions; PAT: Paternal.
Figure modified from [Citation1] Creative Commons Attribution License 4.0 (CC BY).
![Figure 5. Transcription across a paternal germline differentially methylated region facilitates methylation imprint establishment in the male germline. (A) Broad, low-level transcription runs across a PAT gDMR and facilitates de novo DNA methylation of the PAT gDMR [Citation1] by DNMT3A. (B) A typical PAT gDMR is methylated in the chromosome inherited from the sperm (bottom). The same sequence is unmethylated and functions (in this case, as an enhancer-insulator bound by CTCF, yellow) exclusively in the maternal chromosome (top), affecting imprinted genes of the domain. (C) Truncation of the broad, low-level transcript results in hypomethylated PAT* gDMR. (D) When a hypomethylated PAT* gDMR is inherited from the male germline, imprinted gene expression is absent in the soma.gDMR: Germline differentially methylated regions; PAT: Paternal.Figure modified from [Citation1] Creative Commons Attribution License 4.0 (CC BY).](/cms/asset/55e0e8bb-62ad-4221-b41b-a18d49d147fb/iepi_a_12366747_f0005.jpg)
