Figures & data
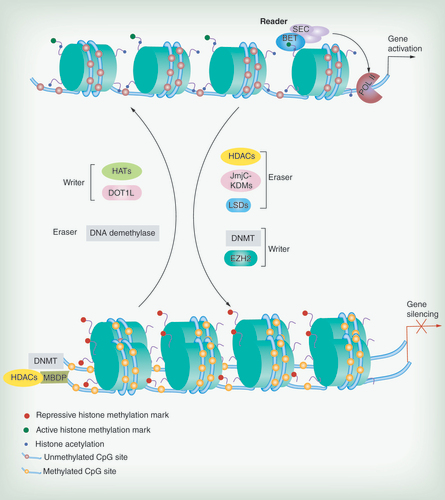
BET, SEC: representative reader (proteins that bind modifications and facilitate epigenetic effects); HATs, DOT1L, DNMT, EZH2: representative writers (enzymes that establish DNA methylation or histone modifications); histone deacetylases, JmjC–KDMs, LSDs, DNA demethylase: representative erasers (proteins that remove DNA methylation or histone modification marks).