Figures & data
Table I. Agonist competition binding parameters of muscarinic receptors in tracheal smooth muscle plasma membranes.
Table II. Antagonist competition binding parameters of muscarinic receptor in tracheal smooth muscle plasma membranes.
Table III. Drugs competition binding parameters of muscarinic receptors in 4-DAMP alkylated plasma membranes from tracheal smooth muscle.
Figure 1. Effect of cGMP in the presence or absence of ATP on the [3H]QNB binding from of plasma membranes from BTSM. Binding experiments were carried out in the presence of cGMP (○) and cGMP plus 5 mM ATP (•) and 1,500 pM [3H]QNB, 5 mM MgCl2 and 2–4 µg of membrane proteins were assayed at 37°C as described in Methods. Specific [3H]QNB binding expressed as percentage of binding in the absence of nucleotides. The binding activity in the absence of cGMP was 1,100 ± 130 fmoles/mg protein and 1,450 ± 170 fmoles/mg protein in the presence of cGMP plus ATP. Each point represents the mean ± SE of four different membrane preparations assayed in triplicate.
![Figure 1. Effect of cGMP in the presence or absence of ATP on the [3H]QNB binding from of plasma membranes from BTSM. Binding experiments were carried out in the presence of cGMP (○) and cGMP plus 5 mM ATP (•) and 1,500 pM [3H]QNB, 5 mM MgCl2 and 2–4 µg of membrane proteins were assayed at 37°C as described in Methods. Specific [3H]QNB binding expressed as percentage of binding in the absence of nucleotides. The binding activity in the absence of cGMP was 1,100 ± 130 fmoles/mg protein and 1,450 ± 170 fmoles/mg protein in the presence of cGMP plus ATP. Each point represents the mean ± SE of four different membrane preparations assayed in triplicate.](/cms/asset/6e37e1f3-9b6a-4356-960b-007afc9da4d1/imbc_a_851419_f0001_b.jpg)
Figure 2. Effect of PDE inhibitors on [3H]QNB binding in of plasma membranes from BTSM. Binding experiments were carried out at 37°C as described in Methods in the presence of 2–4 µg of membrane proteins, 1,500 pM [3H]QNB, 5 mM MgCl2, 5 mM ATP and (•) increasing concentrations of cGMP or (▪) the specific inhibitor of PDE V, zaprinast (100 nM) or (○) the non-specific inhibitor of PDEs, IBMX (10 μM). Each point represents the mean ± SE of four different membrane preparations assayed in triplicate.
![Figure 2. Effect of PDE inhibitors on [3H]QNB binding in of plasma membranes from BTSM. Binding experiments were carried out at 37°C as described in Methods in the presence of 2–4 µg of membrane proteins, 1,500 pM [3H]QNB, 5 mM MgCl2, 5 mM ATP and (•) increasing concentrations of cGMP or (▪) the specific inhibitor of PDE V, zaprinast (100 nM) or (○) the non-specific inhibitor of PDEs, IBMX (10 μM). Each point represents the mean ± SE of four different membrane preparations assayed in triplicate.](/cms/asset/abcdd747-de97-440f-81cc-5e2f0de0a72d/imbc_a_851419_f0002_b.jpg)
Figure 3. Kinetic curves of the binding of [3H]QNB in of plasma membranes from BTSM. Saturation concentration of [3H]QNB was 1,500 pM, 5 mM MgCl2 and 2–4 µg of membrane proteins were assayed in the presence of IBMX (10 μM) at 37°C as described in Methods. Binding experiments were carried out in the presence of 10 nM cGMP (○), 5 mM ATP (▪) and 10 nM cGMP plus 5 mM ATP (•). Specific [3H]QNB binding expressed as fmoles/mg protein. Each point represents the mean ± SE of four different membrane preparations assayed by triplicate. *Statistical significant (p < 0.05). Insert shows the plot of the best line of fit through the first half-life of each curve from Figure 3, which is a method used to calculate the association rate constants (kon) as described (Bennett and Yamamura Citation1985).
![Figure 3. Kinetic curves of the binding of [3H]QNB in of plasma membranes from BTSM. Saturation concentration of [3H]QNB was 1,500 pM, 5 mM MgCl2 and 2–4 µg of membrane proteins were assayed in the presence of IBMX (10 μM) at 37°C as described in Methods. Binding experiments were carried out in the presence of 10 nM cGMP (○), 5 mM ATP (▪) and 10 nM cGMP plus 5 mM ATP (•). Specific [3H]QNB binding expressed as fmoles/mg protein. Each point represents the mean ± SE of four different membrane preparations assayed by triplicate. *Statistical significant (p < 0.05). Insert shows the plot of the best line of fit through the first half-life of each curve from Figure 3, which is a method used to calculate the association rate constants (kon) as described (Bennett and Yamamura Citation1985).](/cms/asset/c5722bd0-bb93-4438-bd04-fff6d6788505/imbc_a_851419_f0003_b.jpg)
Table IV. Effects of nucleotides on the [3H]QNB binding parameters in tracheal smooth muscle plasma membranes.
Table V. Effect of cGMP in the presence of ATP on the agonist and antagonist competition binding parameters of muscarinic receptors in tracheal smooth muscle plasma membranes.
Figure 4. Effect of cGMP on the [3H]QNB binding in Control plasma membranes and 4-DAMP mustard-alkylated plasma membranes. The [3H]QNB binding curves were perrformed as described in Methods for both native membranes (•) and 4-DAMP mustard-alkylated membranes (○) in the presence of 5 mM ATP, 5 mM MgCl2 and increasing cGMP concentrations are indicated. Specific [3H]QNB binding is expressed as percentage of binding in the absence of nucleotides. Binding experiments were carried out at 1,500 pM [3H]QNB and 2–4 µg of membrane proteins were assayed at 37°C. The total binding activity was 1,570 ± 170 fmoles/mg protein in Control membranes and 549 ± 20 fmoles/mg protein in 4-DAMP mustard alkylated-membranes. Each point represents the mean ± SE of four different membrane preparations assayed in triplicate.
![Figure 4. Effect of cGMP on the [3H]QNB binding in Control plasma membranes and 4-DAMP mustard-alkylated plasma membranes. The [3H]QNB binding curves were perrformed as described in Methods for both native membranes (•) and 4-DAMP mustard-alkylated membranes (○) in the presence of 5 mM ATP, 5 mM MgCl2 and increasing cGMP concentrations are indicated. Specific [3H]QNB binding is expressed as percentage of binding in the absence of nucleotides. Binding experiments were carried out at 1,500 pM [3H]QNB and 2–4 µg of membrane proteins were assayed at 37°C. The total binding activity was 1,570 ± 170 fmoles/mg protein in Control membranes and 549 ± 20 fmoles/mg protein in 4-DAMP mustard alkylated-membranes. Each point represents the mean ± SE of four different membrane preparations assayed in triplicate.](/cms/asset/bb817038-9193-4603-9c67-b33ad78d9313/imbc_a_851419_f0004_b.jpg)
Figure 5. (A) Effect of cyclic nucleotide analogs (activator of PKG on the muscarinic activity of plasma membranes from BTSM. [3H]QNB binding in plasma membranes from BTSM was performed using 1,500 pM [3H]QNB and 2–3 µg of membrane proteins were assayed at 37°C as described in Methods. A titration of cGMP was performed in the presence of 5 mM ATP and 5 mM MgCl2 under increasing concentration of Sp-8-pCPT-cGMPS were 0 (▪), 0.25 (•), 0.5 (□), 1.0 (○) and 5.0 μM (◊), respectively. (Insert: Activation curve obtained by plotting binding data obtained at 1 nM cGMP). (B) Effect of cyclic nucleotide analogs (inhibitor) of PKG on the muscarinic activity of plasma membranes from BTSM. [3H]QNB binding in plasma membranes from BTSM was performed using 1,500 pM [3H]QNB and 2–3 µg of membrane proteins were assayed at 37°C as described in Methods. A titration of cGMP was performed in the presence of 5 mM ATP and 5 mM MgCl2 under increasing concentration of Rp-8-pCPT-cGMPS (Insert: Inhibition curve obtained by plotting binding data obtained at 10 nM cGMP). Sp-8-pCPT-cGMPS and Rp-8-pCPT-cGMPS concentrations were 0 (▪), 0.25 (•), 0.5 (□), 1.0 (○) and 5.0 μM (◊), respectively. Each point represents the mean ± SE of four different plasma membrane preparations assayed in triplicate.
![Figure 5. (A) Effect of cyclic nucleotide analogs (activator of PKG on the muscarinic activity of plasma membranes from BTSM. [3H]QNB binding in plasma membranes from BTSM was performed using 1,500 pM [3H]QNB and 2–3 µg of membrane proteins were assayed at 37°C as described in Methods. A titration of cGMP was performed in the presence of 5 mM ATP and 5 mM MgCl2 under increasing concentration of Sp-8-pCPT-cGMPS were 0 (▪), 0.25 (•), 0.5 (□), 1.0 (○) and 5.0 μM (◊), respectively. (Insert: Activation curve obtained by plotting binding data obtained at 1 nM cGMP). (B) Effect of cyclic nucleotide analogs (inhibitor) of PKG on the muscarinic activity of plasma membranes from BTSM. [3H]QNB binding in plasma membranes from BTSM was performed using 1,500 pM [3H]QNB and 2–3 µg of membrane proteins were assayed at 37°C as described in Methods. A titration of cGMP was performed in the presence of 5 mM ATP and 5 mM MgCl2 under increasing concentration of Rp-8-pCPT-cGMPS (Insert: Inhibition curve obtained by plotting binding data obtained at 10 nM cGMP). Sp-8-pCPT-cGMPS and Rp-8-pCPT-cGMPS concentrations were 0 (▪), 0.25 (•), 0.5 (□), 1.0 (○) and 5.0 μM (◊), respectively. Each point represents the mean ± SE of four different plasma membrane preparations assayed in triplicate.](/cms/asset/085e947b-c387-4f5f-9146-a3911cf99089/imbc_a_851419_f0005_b.jpg)
Figure 6. Western blotting for PKG-II in P1 plasma membranes fractions from BTSM. Plasma membrane fraction (P1) (20 µg) was subjected to 10% PAGE-SDS. Proteins were transfer to PDVF membranes and Western blot analysis with rabbit antisera directed against the PKG-II was performed. In addition, to identify the immunoreactivity proteins (positive bands), a Precision Plus protein™ Western C™ Pack from BioRAD and the enhanced chemiluminescence method as described by the manufacturer (Amersham ECL Western Blotting Reagent Pack from GE/Healthcare, Life Sciences) were used to identify the positive bands.

Figure 7. Autoradiograms of m2 and m3-immunoprecipitated 32P-labeled polypeptides. Plasma membranes fraction was subjected to [32P]phosphorylation using [γ32P]ATP as described in Methods. Later, [32P]-labeled membranes were solubilized using a detergent mixture containing 0.1% Digitonin-0.02% sodium cholate and incubated with specific anti m3 or anti m2AChR antibodies as described in Methods. Immunoprecipitates were collected with protein A/G-agarose beads by low speed centrifugation an incubated with cracking SDS solution and subjected to 10% SDS-PAGE. Gels were dried and exposed during 7–12 days at −80°C under X-film (Hyperfilm®). 32P-labeled m3-AChR immunoprecipitated polypeptides in the absence (lane A) or presence (lane B) of 10 nM cGMP and 32P-labeled m2-AChR immunoprecipitated polypeptides in the absence (lane C) or presence (lane D) of 10 nM cGMP. Arrows indicates the molecular masses m3AChR of 67 KDa and 52 KDa for m2AChR. All 32P-labeled phosphoproteins were adjusted to the same level for comparison.
![Figure 7. Autoradiograms of m2 and m3-immunoprecipitated 32P-labeled polypeptides. Plasma membranes fraction was subjected to [32P]phosphorylation using [γ32P]ATP as described in Methods. Later, [32P]-labeled membranes were solubilized using a detergent mixture containing 0.1% Digitonin-0.02% sodium cholate and incubated with specific anti m3 or anti m2AChR antibodies as described in Methods. Immunoprecipitates were collected with protein A/G-agarose beads by low speed centrifugation an incubated with cracking SDS solution and subjected to 10% SDS-PAGE. Gels were dried and exposed during 7–12 days at −80°C under X-film (Hyperfilm®). 32P-labeled m3-AChR immunoprecipitated polypeptides in the absence (lane A) or presence (lane B) of 10 nM cGMP and 32P-labeled m2-AChR immunoprecipitated polypeptides in the absence (lane C) or presence (lane D) of 10 nM cGMP. Arrows indicates the molecular masses m3AChR of 67 KDa and 52 KDa for m2AChR. All 32P-labeled phosphoproteins were adjusted to the same level for comparison.](/cms/asset/9cf33a58-1f37-4774-b923-299c69788ca8/imbc_a_851419_f0007_b.jpg)
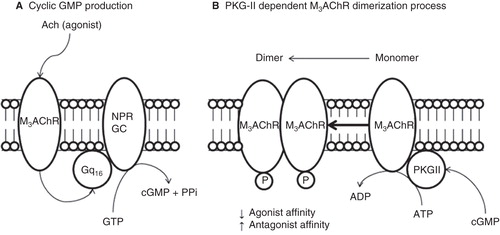
Figure 8. Proposed model of molecular regulation of M3AChR induced by PKG-II phosphorylation at plasma membrane from BTSM. All molecular compnents are membrane bound entities. (A) Muscarinic agonist (Acetylcholine) binds to M3AChR and activates the heterotrimeric Gq16, which stimulates the NPR-GC and increases the cGMP production. (B) Cyclic GMP activates the PKG-II and produces M3AChR phosphorylation inducing a conformation changes to produce M3AChR dimer, which stabilizes or ‘freezes' the M3AChR population, in a refractory state to agonist activation, and prone to antagonist binding, which helps to understand the molecular mechanisms of muscarinic antagonist drug action.