Figures & data
Figure 1. Simple mechanisms of pumps and channels. (A) Alternating access or ‘flip-flop’ model of pump function. A conformational change converts the MSDs between ‘outward facing’ and ‘inward facing’ orientations, alternatively exposing the substrate (red) to one side of the membrane or the other. (B) Single gate model of channel function. A localized conformational change allows the channel to exist in ‘closed’ (transport not allowed) or ‘open’ (transport allowed) states. (C) Two gate model of pump function. Two gates open or occlude the transport pathway. In order for active transport to occur, both gates are never open simultaneously, since this would create a passive, open ion channel-type transport pathway. As a result, it is presumed that a substrate occluded state (centre) must exist. This is a greatly simplified version; in order for active transport to occur, a cyclical model with at least one effectively irreversible step must exist.
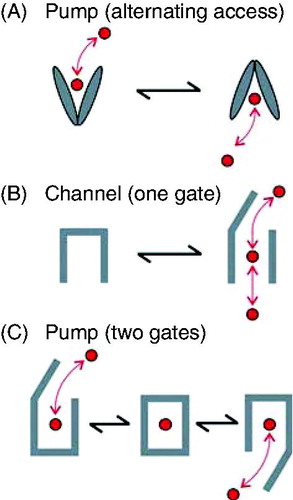
Figure 2. Structures of example ABC proteins. Published structures of Sav1866 (NBDs associated, MSDs outward facing), P-glycoprotein (NBDs dissociated, MSDs inward facing), TM287/288 (NBDs partially associated, MSDs inward facing) and ABCB10 (NBDs associated, MSDs inward facing). Protein Data Bank (PDB) accession numbers for these structures are 2ONJ, 3G5U, 3QF4, and 4AYT respectively. The Figure was prepared using PyMol software (Schrödinger, LLC, Portland, OR, USA).
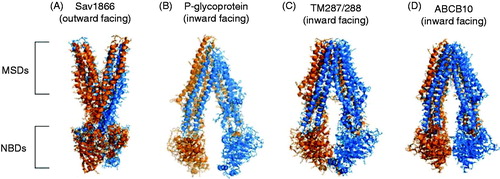
Figure 3. Examples of CFTR homology models based on ABC protein templates. (A) Outward facing model, based on Sav1866 template, published by Serohijos et al. (Citation2008); a very similar model was also published by Mornon et al. (Citation2008). (B) Inward facing model based on MsbA template, published by Mornon et al. (Citation2009). (C) ‘Channel-like’ configuration, based on Sav1866 template, published by Dalton et al. (Citation2012). In each case, the N-terminal ‘half’ of CFTR (TMs 1-6, NBD2) is coloured blue, and the C-terminal half (TMs 7-12, NBD2) orange. The R domain in (A) and (B) is coloured gray; the R domain was not included in the model shown in (C). Each panel was visualized with PyMol using coordinates provided in the described publications.
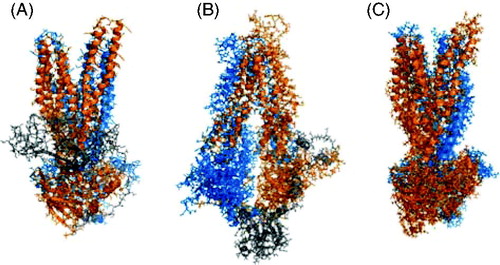
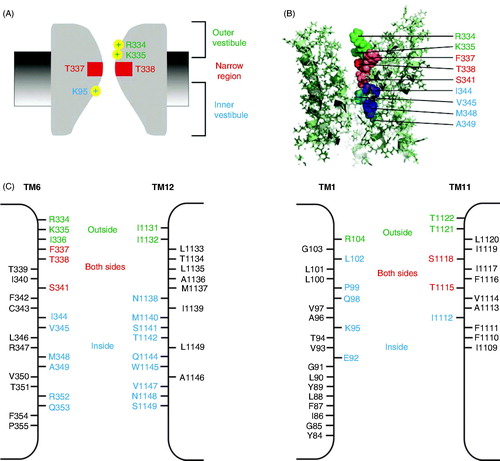
Figure 4. Proposed arrangement of TMs in CFTR-MSDs. (A, B) Symmetrical arrangement of TMs in the ‘channel-like’ homology model presented by Dalton et al. (Citation2012), viewed from the side (A) or from the extracellular side of the membrane (B). Symmetrical ‘pairs’ of TMs from the N- and C-terminal parts of the protein are indicated in the same colour, namely green (TMs 1, 7), brown (TMs 2, 8), yellow (TMs 3, 9), gray (TMs 4, 10), blue (TMs 5, 11) and red (TMs 6, 12). (C) Asymmetrical arrangement of pore-lining TMs 1, 6, 11 and 12. The apparent location of the pore lumen is indicated by the indicated side chains of pore-lining residues from approximately the same level in each of these four TMs, namely K95 (TM1), I344 (TM6), I1112 (TM11) and S1141 (TM12) (see also ).
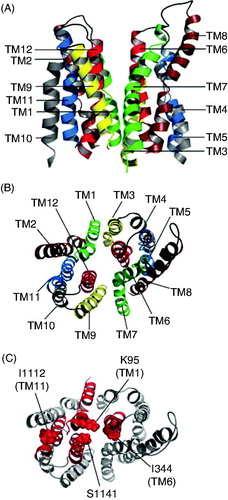
Figure 5. Working model of different regions of the open channel pore. (A) Functional cartoon model of the pore, summarizing key features described in the text: A narrow central pore region lined by TM6 residues F337 and T338 (red), a shallow outer vestibule including positively charged TM6 residues R334 and K335 (green), and a deeper, wide inner vestibule including positively charged TM1 residue K95 (blue). (B) Approximate location of pore-lining TM6 side chains proposed to contribute to these three functionally separable pore regions, according to the same colour scheme. The image shows a cross-section through the MSD region of the model presented by Dalton et al. (Citation2012) (see also ). (C) Pore-lining side chains identified in structurally symmetrical TMs 6 and 12 (left) and structurally asymmetric TMs 1 and 11 (right). Based on SCAM work from the author’s laboratory (El Hiani and Linsdell, Citation2010, Qian et al. Citation2011, Wang et al. Citation2011, Citation2014), residues that (when mutated to cysteine) are accessible to large cysteine-reactive from the outside are coloured green, those accessible from the inside are coloured blue, and those accessible from both sides of the membrane are coloured red, a colour scheme that is deliberately common with that used in (A) and (B). Apparently non-pore-lining side chains are in black. Broadly similar results were obtained by other groups (Alexander et al. Citation2009, Bai et al. Citation2010, Citation2011, Gao et al. Citation2013), although importantly these other groups did not find that any residues were accessible from both sides of the membrane. Nevertheless, the ‘cut-off’ region that prevents permeation of large cysteine-reactive reagents from one side of the membrane to the other, which most likely corresponds to the narrowest part of the open channel pore, is consistently observed (see text).