Figures & data
Table 1. Primer sets for MSP analysis.
Figure 1. Effects of FB1 (1–200 µM) on cell viability and cytotoxicity by XTT (a) and LDH tests (b) in Clone 9 and NRK-52E cells after 24 h incubation. Data are presented as mean ± standard deviation of at least three determinations. *Different from control at p < 0.001.
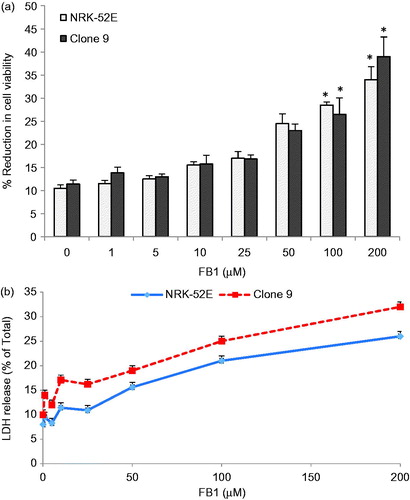
Figure 2. A representative HPLC chromatogram of all five deoxyribonucleosides in standard mixture (a) and in hydrolyzed DNA of NRK-52E cells (b). All deoxyribonucleosides were well separated based on established retention times with standard chemicals. dC, 2′-deoxycytidine; 5-mdC, 5-methyl-2′-deoxycytidine;2-dT, 2′-deoxythymidine; 2-dG, 2′-deoxyguanosine monohydrate; 2-dA, 2′-deoxyadenosine monohydrate.
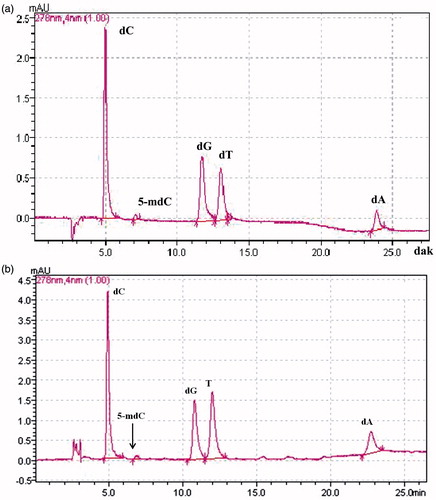
Figure 3. Effects of FB1 (1, 5, 10, 25, and 50 µM) on global DNA methylation in NRK-52E and Clone 9 cells after 24 h incubation. Data are presented as mean ± standard deviation (n = 6). Genomic DNA was extracted and hydrolyzed to deoxyribonucleosides. Global methylation status was quantified by HPLC-UV/DAD.
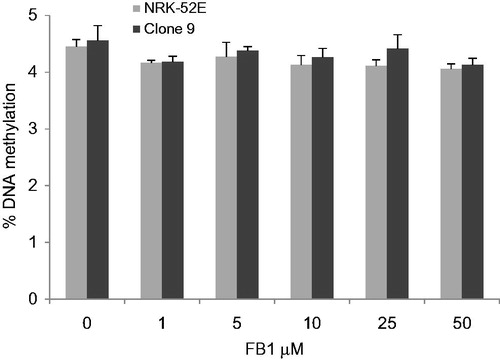
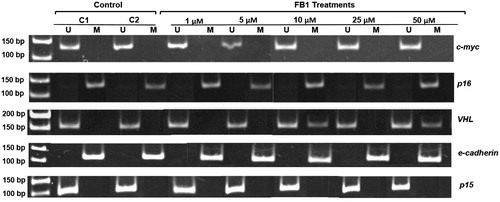
Figure 4. Effect of FB1 on methylation status of c-myc, p16, VHL, e-cadherin, and p15 genes in Clone 9 cells. A representative sample of Clone 9 cells treated with FB1 (1, 5, 10, 25, and 50 µM) for 24 h is shown. Methylation was determined by bisulfite modification of the genomic DNA and MSP using primers for the unmethylated (U) or methylated (M) promoter sequence. C1 and C2 = PBS (1%) as a control instead of FB1 treatment.
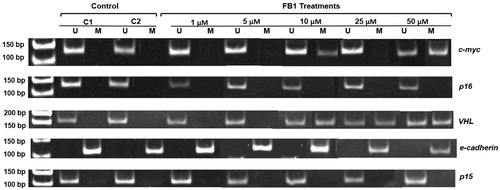
Figure 5. Effect of FB1 on methylation status of c-myc, p16, VHL, e-cadherin, and p15 genes in NRK-52E cells. A representative sample of NRK-52E cells treated with FB1 (1, 5, 10, 25, and 50 µM) for 24 h is shown. Methylation was determined by bisulfite modification of the genomic DNA and MSP using primers for the unmethylated (U) or methylated (M) promoter sequence. C1 and C2 = PBS (1%) as a control instead of FB1 treatment.