Figures & data
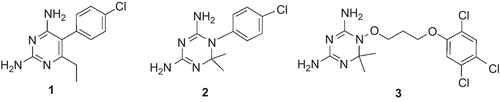
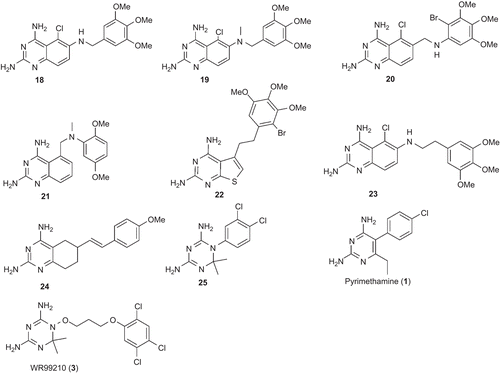
Figure 2. Chemical structures of the training set compounds used for pharmacophore model generation.
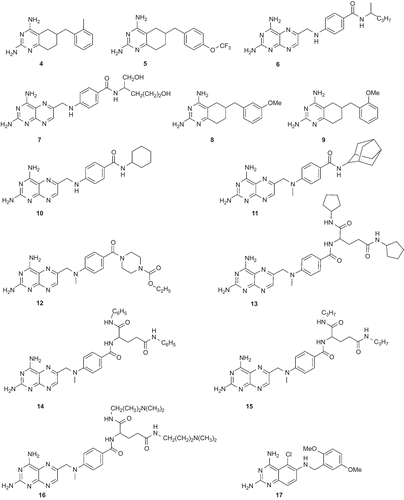
Table 1. Summary of the hypotheses generated by a Catalyst/HipHop run.
Table 2. pIC50, number of conformers, best-fit values, and ΔE values of the training set compounds.
Figure 3. Mapping of 4 (a), 8 (b), 9 (c), 17 (d), 18 (e), 19 (f), and WR99210 (3) (g) onto hypo1. Pharmacophoric features are color-coded (violet, hydrogen bond donor; blue, hydrophobic aliphatic; light blue, aromatic hydrophobic).
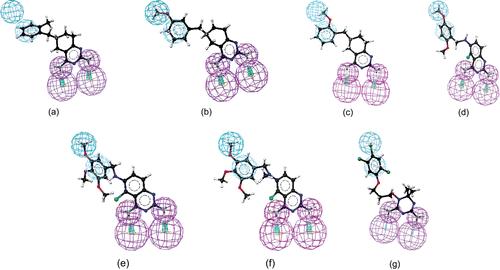
Table 3. Docking scores of hits identified from the virtual screening study.
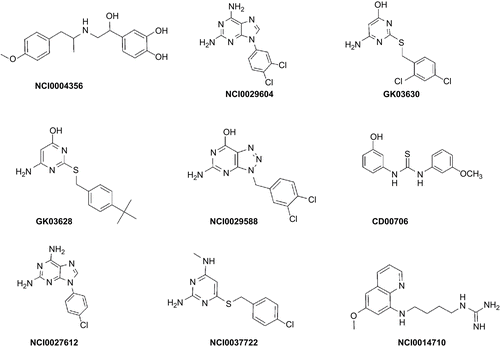
Figure 5. Mapping of NCI00043568 (a), GK-03628 (b), NCI0029588 (c), CD00706 (d), NCI0037722 (e), and NCI0014710 (f) onto hypo1. Pharmacophoric features are color-coded (violet, hydrogen bond donor; blue, hydrophobic aliphatic; light blue, aromatic hydrophobic).
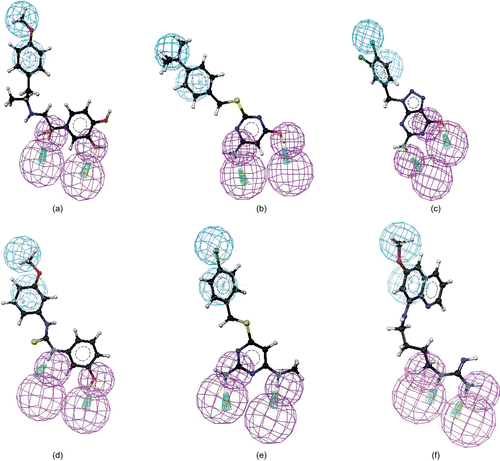
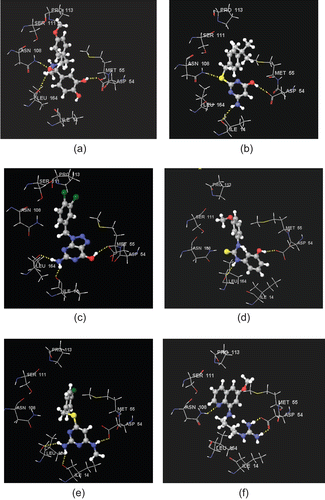
Figure 6. Stereoview of XPGlide predicted binding poses of (a) NCI004356, (b) GK03628, (c) NCI0029588, (d) CD00706, (e) NCI0037722, and (f) NCI0014710 in the active site of the quadruple-mutant PfDHFR enzyme. For the sake of clarity, only important amino acid residues are given.