Figures & data
Figure 1. Chemical structures of the training set compounds together with their experimental Ki values in µM used for pharmacophore generation.
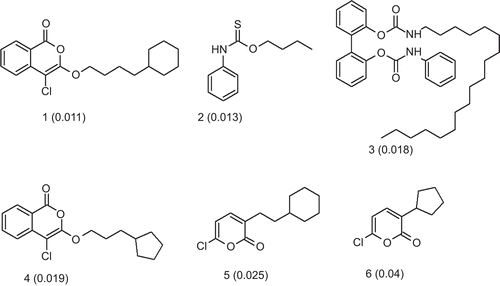
Table 1. Summary of the generated common feature pharmacophore models.
Table 2. Best fit values and absolute energy for the training set compounds respective to “Hypo 1” and “Hypo 2”.
Figure 2. (A) Three-dimensional spatial arrangement of the best pharmacophore hypothesis “Hypo 1”. (B) The distance constraints between the chemical features. Green colour represents hydrogen bond acceptor (HBA) and cyan colour represents hydrophobic (HY) features.
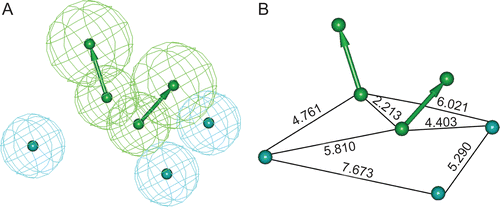
Table 3. Statistical parameters and scores from decoy set validation for “Hypo 1”.
Figure 3. Compound 1 (A) and compound 6 (B) in the training set mapped on “Hypo 1”. Green colour represents hydrogen bond acceptor (HBA) and cyan colour represents hydrophobic (HY) features.
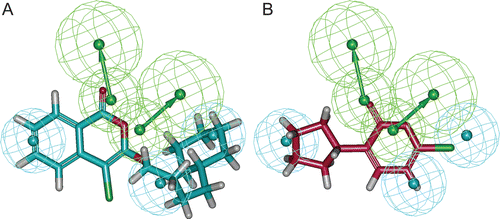
Figure 4. Pharmacophore mapping of final database hit compounds on the best pharmacophore hypothesis “Hypo 1”, (A) SEW00846 represented in green colour, (B) NCI0040784 represented in violet colour, (C) GK03167 represented in blue colour, and (D) CD10645 represented in cyan colour. In the pharmacophore hypothesis, green represents hydrogen bond acceptor (HBA) and cyan represents hydrophobic (HY) features.
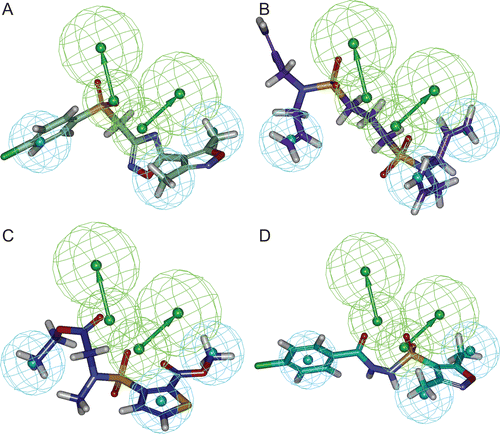
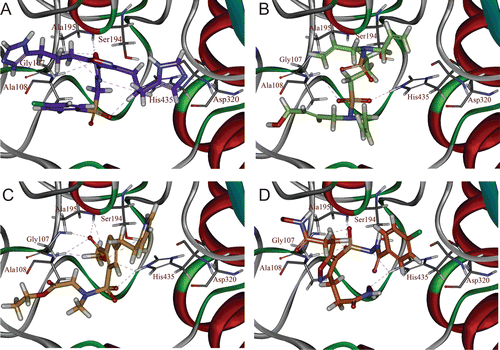
Figure 5. Binding orientations of database hit compounds: (A) SEW00846, (B) NCI0040784, (C) GK03167, and (D) CD10645 are shown in cyan, red, blue, and magenta colours, respectively. Hydrogen bonds are shown in dotted lines.
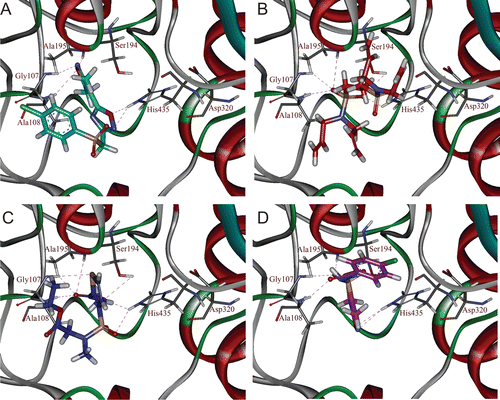
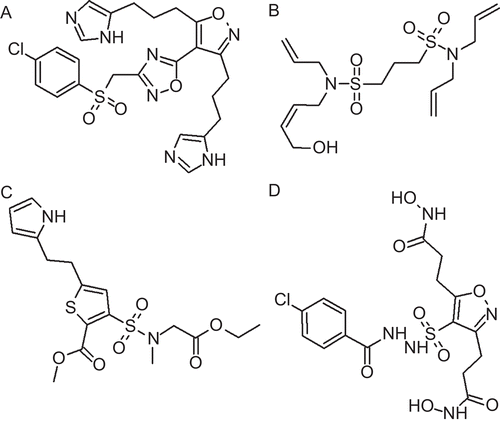
Figure 6. 2D representation of the database hit compounds: (A) SEW00846, (B) NCI0040784, (C) GK03167, and (D) CD10645.
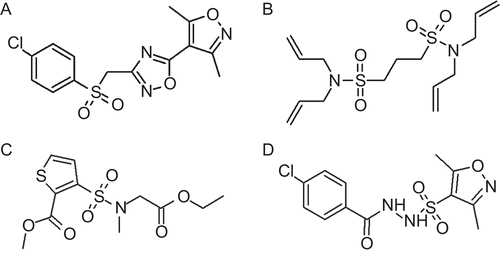
Table 4. GOLD scores, AutoDock binding energies, and SYLVIA synthetic accessibility scores of top 10 optimized compounds.