Figures & data
Figure 1. Chemical structure of the LMW mushroom compounds with anti-cancer potential isolated from mushrooms.
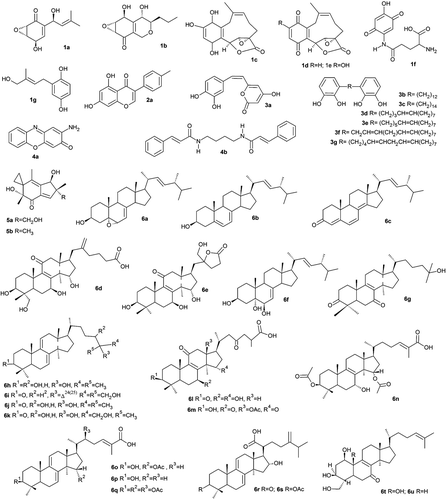
Table 1. Re-docking and cross-docking results using the selected Mdm2 crystal structures.
Figure 2. Docking conformation of three known Mdm2 inhibitors. (a) Representation of the Mdm2 structure with the co-crystallized benzodiazepine inhibitor (PDB:1T4E). Superimposition of crystal (sticks and balls representation, green) and docked conformations (wire representation, yellow) for: (a) the benzodiazepine derivative, (b) the imidazo-indole derivative and (c) the chromenotriazolopyridine derivative. Superimpositions obtained by aligning the three Mdm2 structures using Pymol.
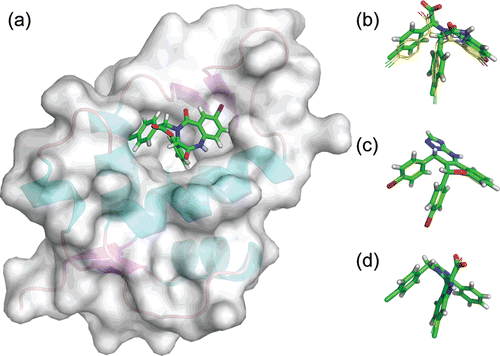
Table 2. Virtual screening of the LMW mushroom compound database using AutoDock4.
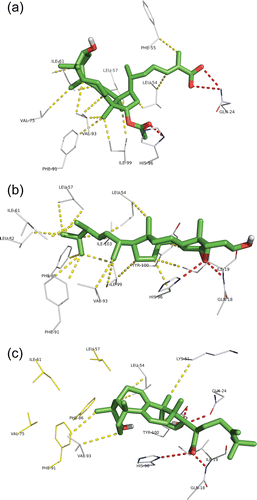
Figure 3. Docking conformation of the top ranked LMW mushroom compounds in the Mdm2 interaction site: (a) ganoderic acid X, (b) 5,6-epoxy-24(R)-methylcholesta-7,22-dien-3β-ol and (c) polyporenic acid C. All the compounds are represented in green sticks and balls representation. The Mdm2 residues from the interaction site interacting with the compounds are represented in white wire representation. Hydrogen bonds are represented in traced red (bond distances between 2.8 and 3.2 Å), hydrophobic interactions in traced yellow (bond distances between 3.5 and 4 Å).