Figures & data
Figure 1. Structure of fukugetin. IUPAC name: 8-[(2S,3R)-5,7-dihydroxy-2-(4-hydroxyphenyl)-4-oxo-2, 3-dihydrochromen-3-yl]-2-(3,4-dihydroxyphenyl)-5,7-dihydroxychromen-4-one.
![Figure 1. Structure of fukugetin. IUPAC name: 8-[(2S,3R)-5,7-dihydroxy-2-(4-hydroxyphenyl)-4-oxo-2, 3-dihydrochromen-3-yl]-2-(3,4-dihydroxyphenyl)-5,7-dihydroxychromen-4-one.](/cms/asset/62426d34-6703-4ffa-a757-a68c5d6e6dd5/ienz_a_668539_f0001_b.gif)
Table 1. Crystal data and structure refinement for fukugetin.
Figure 2. ORTEP view of fukugetin (S-enantiomers) showing the arbitrary atom labelling. Ellipsoids represent 50% probability level.
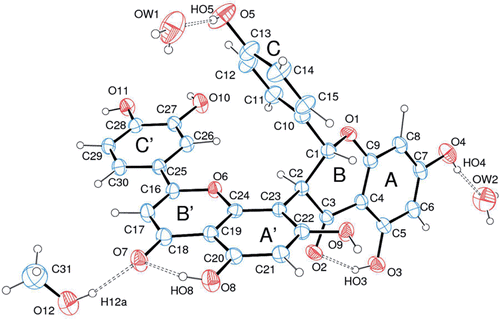
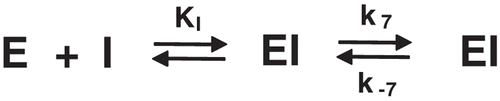
Figure 3. Slow-binding inhibition of papain (upper) and cruzain (lower) by fukugetin. Left− time course of inactivation. Right− first-order constant (Kobs) as function of inhibitor concentration.
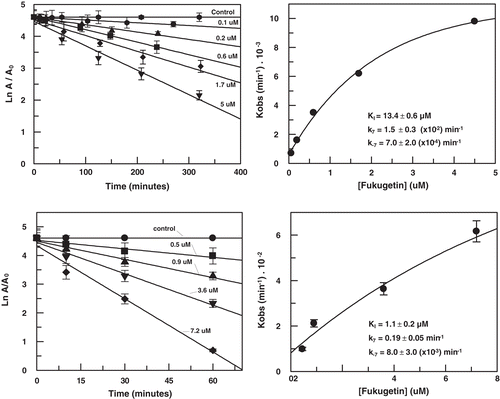
Table 2. Kinetic parameters from papain and cruzain inhibition by fukugetin (means ± standard deviation).
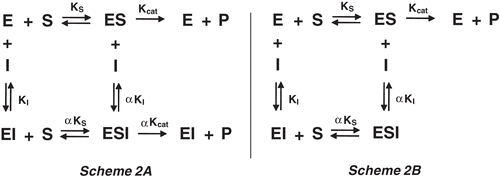
Figure 4. (A) Lineweaver–Burk plot for hydrolysis of Cbz-Phe-Arg-MCA by papain in the presence and absence of fukugetin and (B) the secondary plots of showing the hyperbolic inhibition type mixed on fukugetin concentration. The reaction was carried out in the absence (• ) and in the presence of fukugetin; Δ, 1.5µM; ▪, 3µM; ◊, 6µM and ◆, 12µM.
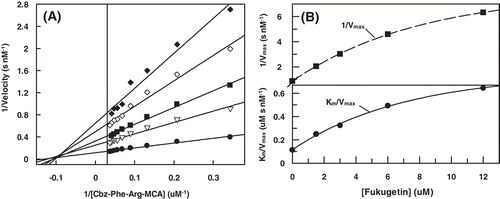
Figure 5. (A) Lineweaver–Burk plot for hydrolysis of Cbz-Phe-Arg-MCA by cruzain in the presence and absence of fukugetin and (B) the secondary plots showing the dependence of the axis 1/Vmax and the Km/Vmax on the fukugetin concentration. The reaction was carried out in the absence (• ) and in the presence of fukugetin; □ 1µM; ▴, 2µM; ◊, 5µM; ◆, 10µM and ◊, 18µM.
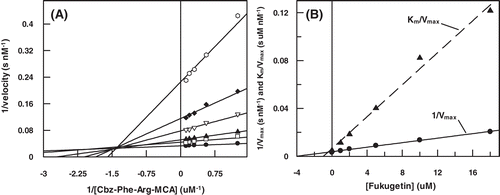
Figure 6. Molecular docking of papain (left) and cruzain (right) fukugetin inhibitor. The proteins are represented based on electrostatic potential surface (EPS); inhibitor carbons are pink, hydrogens are white and oxygens are red. The papain and cruzain structure files were downloaded from the Protein Data Bank (PDB code 1PE6 and 1F2C, respectively).
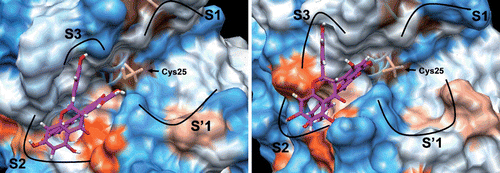
Figure 7. Best docking poses for fukugetin inside the subsites of cruzain (left) and papain (right). The hydrogen bonds that contribute for inhibitor anchoring are shown as yellow lines and the hydrophobics interactions as green lines. Some amino acids were highlighted due to their key roles in the binding of inhibitor. H atoms in C-H bonds are omitted.
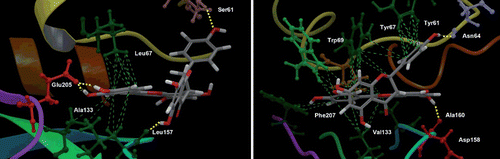
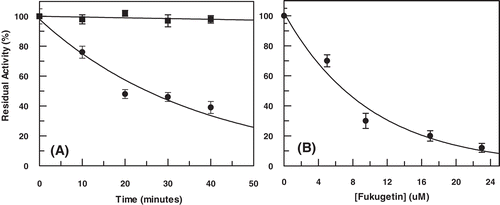
Figure 8. (A) Time course of enzymatic activity from Trypanosoma cruzi extract in the presence of fukugetin 10 µM and (B) enzymatic activity as function of fukugetin concentration. Both experiments were carried out using Cbz-Phe-Arg-MCA 10 µM as substrate.