Figures & data
Scheme 1. Fundamental representation of paraoxonase 1 (PON1) structure. Cys-284 residue is considered to provide the antioxidant effects at the three-dimensional structure of PON1 enzyme.
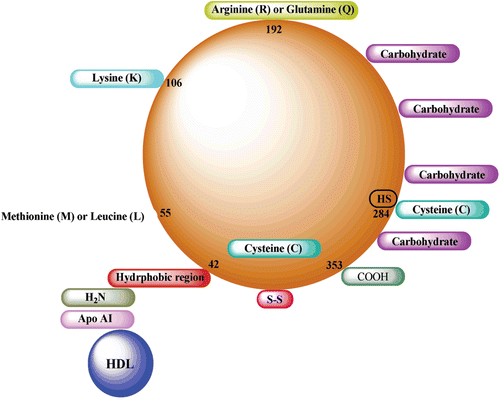
Figure 1. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis analysis of purified paraoxonase 1 (PON1). Lane 1: standard proteins (kD): aldolase (47,500), triosephosphateisomerase (32,000), soy bean trypsin inhibitor (24,000) Da. Lane 2: PON1 from human serum via Sephadex G-200 gel filtration chromatography.
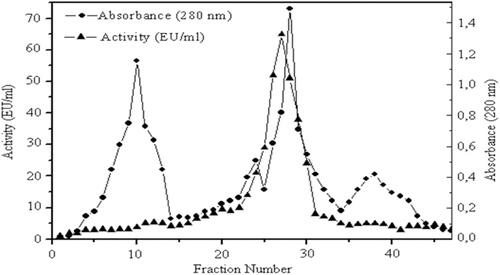
Table 1. Purification steps of PON1 enzyme from human serum by ammonium sulfate precipitation, anion exchange chromatography, and gel filtration chromatography.
Figure 2. Elutiongraph of paraoxonase 1 from human serum by Sephadex G-200 gel filtration chromatography. Fractions from the diethylaminoethyl cellulose-Sephadex column were mixed with glycerol and loaded onto the Sephadex G-200 column (60 cm × 2 cm),which had been equilibrated with 100 mM Na-phosphatebuffer (pH7.0). Elution was performed with the same buffer. Fractions were analyzed for both protein amount (280 nm) and enzyme activity (412 nm).
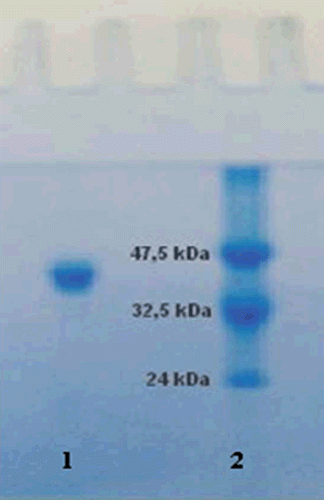
Table 2. Ki values and inhibition types for five different medical drugs.
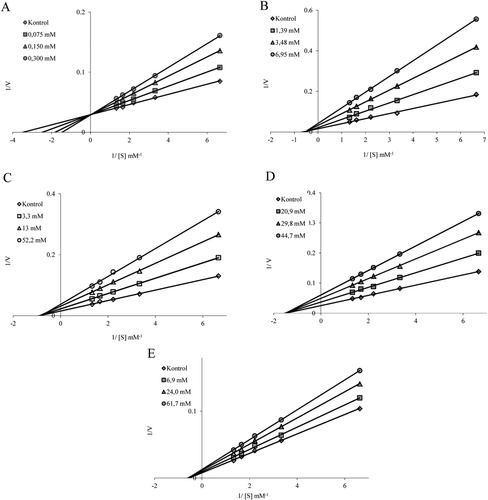
Figure 3. Determination of inhibitiontypesand Ki values of the drugs using Lineweaver–Burk curves. For the determination of Ki values, three different inhibitor concentrations were tested for each drug. Paraoxone was used as substrate at five different concentrations. Control activity was assumed to be 100% in the absence of inhibitor. Activity assays were performed as described in Materials and methods section.