Figures & data
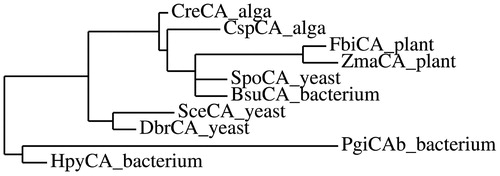
Figure 1. SDS-PAGE of the recombinant PgiCAb purified from E. coli. Lane STD, molecular markers; Lane 1, purified PgiCA from His-tag affinity column in its monomeric and dimeric forms.
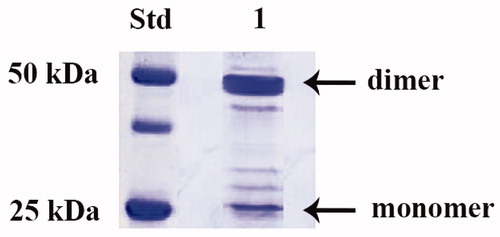
Figure 2. Nucleotide and amino acid sequence of β-CA from P. gingivalis. Amino acid residues participating in the coordination of metal ion are indicated in red, whereas the catalytic dyad involved in the activation of the metal ion coordinated water molecule (Asp97–Arg99) is shown in blue. The asterisk (*) indicates the stop codon.

Figure 3. Multi-alignment of the amino acid sequences of the selected β-CAs from plant, yeast, algae and bacterial organisms. Zinc ligands are indicated in red, catalytic dyad in blue. The asterisk (*) indicates identity at a position; the symbol (:) designates conserved substitutions, while (.) indicates semi-conserved substitutions. Multiple alignment was performed with the program Clustal W, version 2.1. Legend: PgiCAb_bacterium, Porhyromonas gingivalis (Accession number: YP_001929649.1); BsuCA_bacterium, Brucella suis (Accession number: WP_012243428.1); HpyCA_bacterium, Helicobacter pylori (Accession number: BAF34127.1); CspCA_alga, Coccomyxa sp. (Accession number: AAC33484.1); SceCA_yeast, Saccharomyces cerevisiae (Accession number: GAA26059); DbrCA_yeast, Dekkera bruxellensis AWRI1499 (Accession number: EIF49256); SpoCA_yeast, Schizosaccharomyces pombe (Accession number: CAA21790); FbiCA_plant, Flaveria bidentis, isoform I (Accession number: AAA86939.2); ZmaCA_plant; Zea mays (Accession number: NP_001147846.1).

Figure 4. Phylogenetic tree of the β-CAs used in the multi-alignment showed in . The tree was constructed using the program PhyML 3.0. Branch support values are reported at branch points. For the legend see .