Figures & data
Figure 1. Phylogenetic analysis of 16S rDNA similarities of Enterobacter sp. B13 based on the BLAST result using the neighbor-joining method. Scale bar represents 0.01 substitutions per nucleotide position. The organisms and GeneBank accession numbers of analyzed sequences are given in parenthesis.
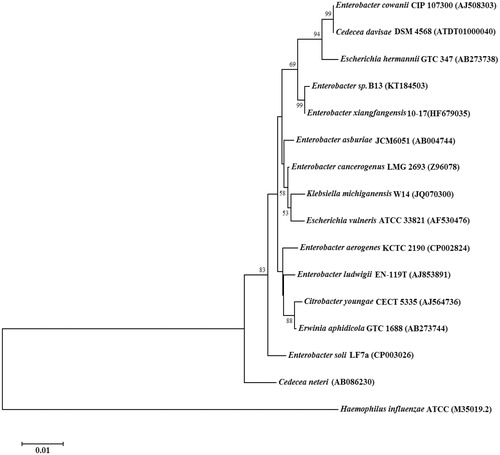
Figure 2. SDS–PAGE analysis (15%) of Enterobacter sp. B13-CA (stained with coomassie brilliant blue). Lane 1, protein molecular weight markers (NEB, P7710S), lane 2, purified Enterobacter sp. B13-CA.
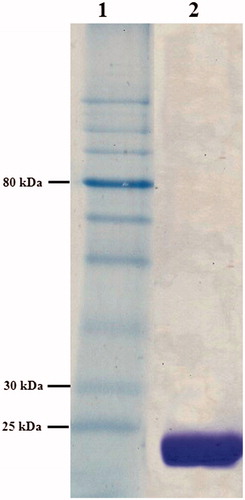
Figure 3. Evolutionary relationship analysis, by the neighbor-joining method, of 21 known CAs from prokaryotes, in comparison to Enterobacter sp. B13-CA (accession numbers are given in parenthesis). Scale bar represents 0.05 substitutions per nucleotide position.
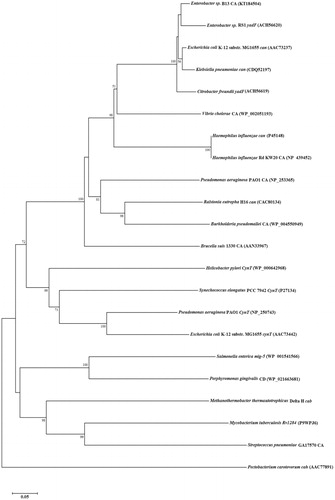
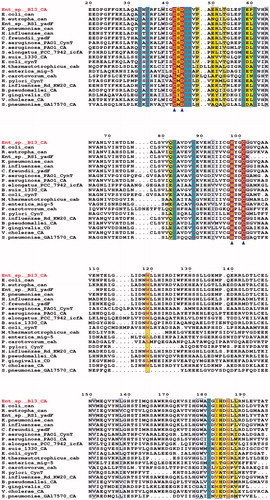
Figure 4. Multiple amino acid sequence alignment of 21 known β-class CAs: Enterobacter sp. B13-CA, E. coli str. K-12 substr. MG1655can, Ralstonia eutropha can, Enterobacter sp. RS1 yadF, Klebsiella pneumoniae can, H. influenzae can, C. freundii yadF, P. aeruginosa PAO1 cynT, P. aeruginosa PAO1 CA, Synechococcus elongates PCC 7942 icfA, B. suis 1330 CA, E. coli str. K-12 substr. MG1655 cynT, M. thermautotrophicus cab, S. enterica mig-5, P. carotovorum cah, H. pylori cynT, H. influenzae Rd KW20 CA, Burkholderia pseudomallei CA, P. gingivalis CD, V. cholerae CA, S. pneumoniae GA17570 CA and M. tuberculosis Rv1284. Triangles indicates the zinc ion-binding residues. Other active side conserved residues were shaded in blue (red letters). The figure was drawn with ESPript.