Figures & data
Fig. 1. Depletion of extracellular vesicles reduces FBS-induced cell migration. Medium was either supplemented with 10% FBS (Medium, non-depleted FBS) or 10% FBS that had been centrifuged for 18 hours to eliminate extracellular vesicle (Medium, EV-depleted FBS). As a control, FBS-free media was used (Plain medium). A549 cells were seeded on one side of a gelatin coated membrane of a Boyden chamber to evaluate cellular transmigration towards the various stimuli on the other side of the membrane. Student's t-test was used to determine significant differences, p<0.05, ***p<0.001. n=3.
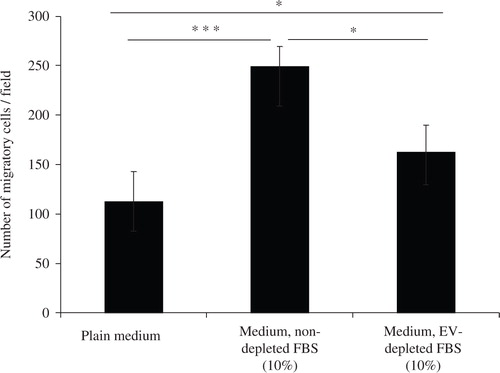
Fig. 2. Characterization of FBS-derived extracellular vesicles. FBS-derived vesicles were collected at 120,000×g for 18 hours, dissolved in PBS and re-pelleted at 120,000×g for 70 minutes. a) To visualize if FBS had vesicular structures present, TEM was performed on the EV-enriched pellet. The scale bar represents 200 nm. b) Immuno-blotting for the exosome enriched proteins; TSG101, CD81, and CD63. Pellet of EVs were obtained from different batch of FBS. Cell lysate from human mast cells (HMC-1) and mouse fibroblast (NIH-3T3), as well as HMC-1 derived exosomes were used as positive controls. One hundred microgram of protein was loaded per well for all samples. c) RNA was isolated using the miRCURY total RNA isolation kit and quantified with a Bioanalyzer. d) The FBS-derived EV-enriched pellet was incubated with RNase A (0.5 µg/µl) alone (i) or was incubated with proteinase K (0.05 µg/µl) prior to the RNase treatment (ii) to study mode of RNA protection. Cellular RNA was used as control for the RNase activity (iii). Graphs are representative of the RNA profile obtained with a Bioanalyzer from two independent experiments. e) PKH67-labelled FBS-derived EVs were incubated with 7×104cells (A549) for 6 or 20 hours and visualized with a fluorescent microscope (40X). PBS was used as control for determining the PKH67 background labelling.
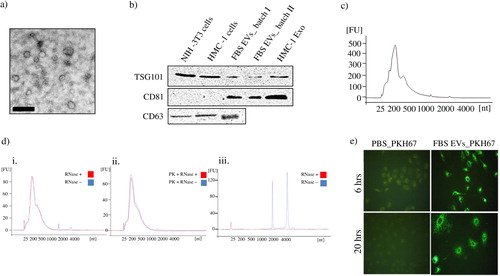
Fig. 3. FBS-derived extracellular vesicles induce migratory phenotype in an epithelial cell line. FBS-derived EVs were collected at 120,000×g for 18 hours, washed in PBS and re-pelleted for 70 minutes at 120,000×g. Isolated vesicles were incubated with A549 cells, but were separated by a gelatin coated membrane in a Boyden chamber to evaluate cellular transmigration towards variable EVs concentrations. FBS-derived EVs showed a dose-dependent response in inducing migration in A549 cells. Kruskal-Wallis test was used to determine significant differences. Comparisons were only made against the control. *p<0.05, **p<0.01, ***p<0.001. n=3. Spearman Rank gave a significant correlation of Rs=0.82.
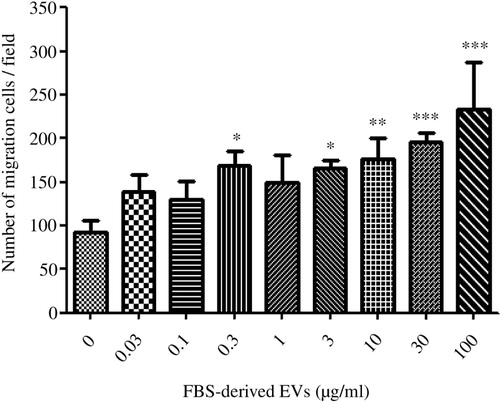
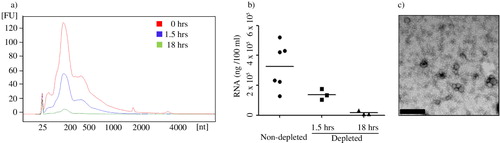
Fig. 4. Analysis of FBS vesicles depletion protocols. EV-depleted FBS was obtained from 1.5 and 18 hours of ultracentrifugation and was used to prepare complete media (10% FBS). The media was never in contact with cells, but used to isolate exosomes from. The RNA was extracted from the 120,000×g pellet and the concentration and nucleotide size was determined by a Bioanalyzer and the Pico chip. a) Data represent the overlay of RNA profile concentration of bovine RNA retained in complete media supernatant and pelleted during exosome isolation. b) Efficiency of exosome depletion with different centrifugation period is compared based on total RNA content in their exosome preparation in complete media. Results represent mean±SEM of sample obtained from 3–6 independent experiments. c) Presence of vesicular structures was determined by transmission electron microscopy. Vesicles are from the pellet preparation from EV-depleted FBS after 18 hours of ultracentrifugation (Bar represents 200 nm).