Figures & data
Figure 1. Jurkat cells captured on antibody Lewis X Clone-1.
(A) Reference diffraction pattern; (B) object diffraction pattern and (C) hologram diffraction pattern; (D) numerical reconstruction of the hologram rendered the 3D image of the cells; (E) segmentation algorithm marked each individual cell; and (F) 3D image of the Jurkat cells.
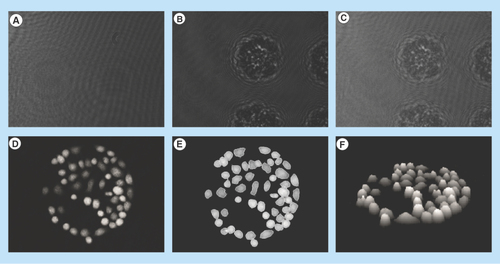
Figure 2. Hologram images of Jurkat cells captured on antibody Lewis X Clone-1.
The images are showing control, DMSO and etoposide-treated cells at four different timepoints: 10, 310, 630 and 950 min. The cells are segmented to analyze cell parameters.
DMSO: Dimethyl sulfoxide.
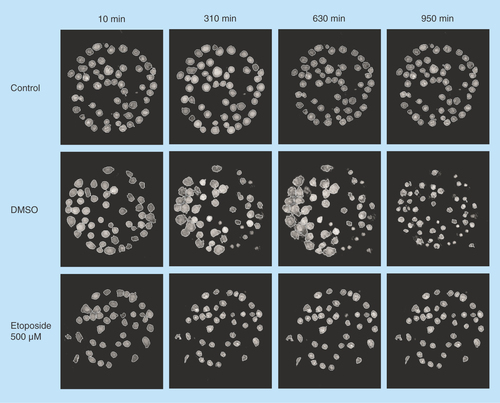
Figure 3. Hologram images of U2932 cells captured on antibody HLA-DR.
The images are showing control, DMSO and etoposide-treated cells at four different timepoints: 10, 310, 630 and 950 min. The cells are segmented to analyze cell parameters.
DMSO: Dimethyl sulfoxide
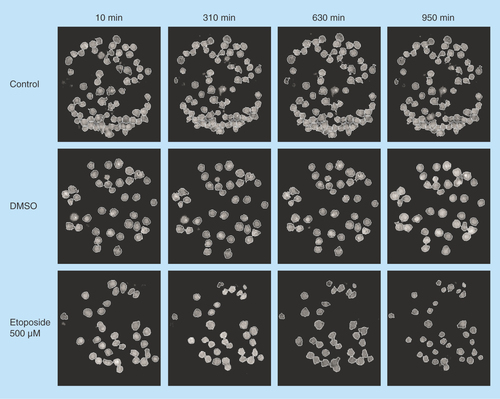
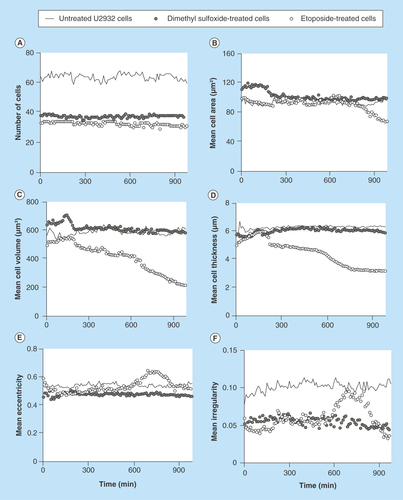
Figure 4. Different cellular parameters of treated (etoposide or dimethyl sulfoxide) or untreated Jurkat cells captured on antibody Lewis X Clone-1 were analyzed over time.
(A) Number of cells; (B) mean cell area; (C) mean cell volume; (D) mean cell thickness; (E) mean cell eccentricity; and (F) mean cell irregularity.
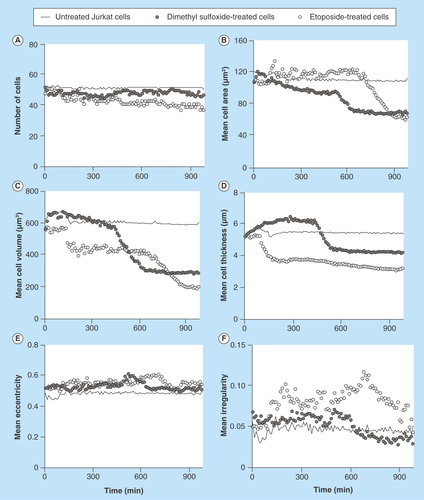
Figure 5. Different cellular parameters of treated (etoposide or dimethyl sulfoxide) or untreated U2932 cells captured on antibody HLA-DR were analyzed over time.
(A) Number of cells; (B) mean cell area; (C) mean cell volume; (D) mean cell thickness; (E) mean cell eccentricity; and (F) mean cell irregularity.