Figures & data
Figure 1. Detection of extended-spectrum β-lactamases-mediated hydrolysis of cefotaxime and ceftazidime by HPLC.
(A & B) Optimization of incubation time for detection of cefotaxime and ceftazidime resistance. Samples were incubated with 100 μg/ml cefotaxime (A) or 20 μg/ml ceftazidime (B) for time periods of 0–12 h. (C & D) Optimization of cefotaxime and ceftazidime concentrations for detection of cefotaxime and ceftazidime resistance. Samples were incubated with 10–100 μg/ml of cefotaxime (C) or 5–50 μg/ml of ceftazidime (D) for 1 h. All experiments were repeated three-times in triplicate. Bars represent the mean proportion of cefotaxime or ceftazidime loss ± standard error of the mean. Incubation times do not include the additional preparation time (∼15 min).
NS: Nonsignificant.
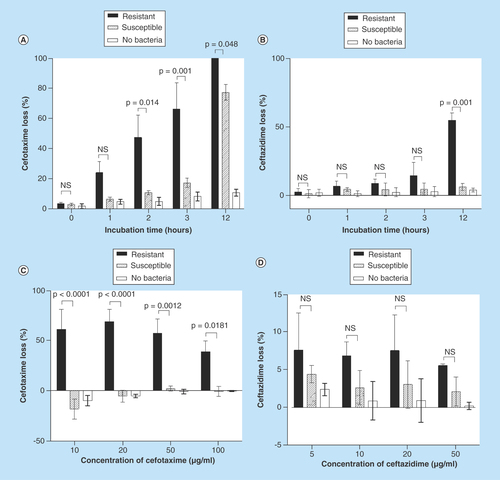
Figure 2. Effect of bacterial concentration on cefotaxime loss.
Various concentrations of cefotaxime-resistant extended-spectrum β-lactamase- and AmpC-producing E. coli (circles) or cefotaxime-susceptible E. coli (triangles) were incubated with 20 μg/ml cefotaxime for 1 h and the loss of cefotaxime determined by the HPLC assay. Each data point is the mean of three technical replicates of assay; four different concentrations (tenfold dilutions) were assessed on four different occasions. The concentration of viable bacterial cells was determined for each experiment.
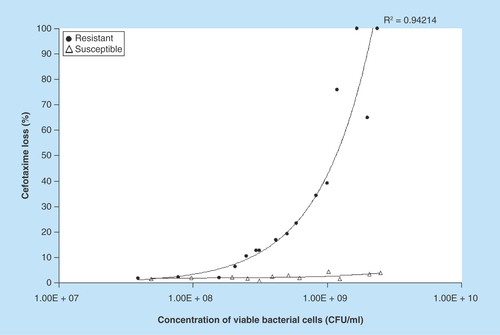
Figure 3. Performance of the HPLC method for the detection of cefotaxime-resistant bacteria.
The receiver operating characteristic curve was calculated to show the sensitivity and specificity of the data from the proportion of cefotaxime loss in cefotaxime-susceptible (n = 27) and cefotaxime-resistant (n = 22) samples. The area under the curve was 0.9302.
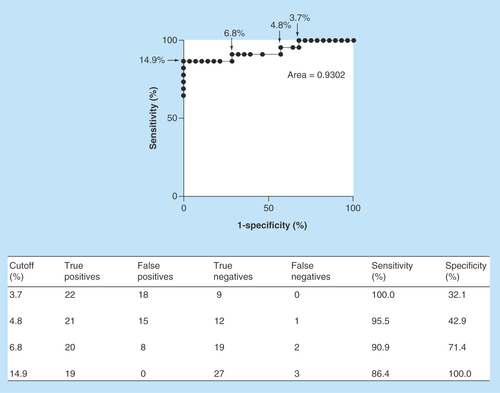
Table 1. Summary of chromatographic parameters for detection of cefotaxime and ceftazidime†.
Table 2. Intra and interday precision and inaccuracy for the quantification of cefotaxime and ceftazidime by HPLC.
