Figures & data
Table 1. Population characteristics of the mothers and infants in the cohort (n = 70)
Figure 1. Cord red blood cell fatty acid measurements in fish oil and control groups. Boxplots represent median and range. Statistical analysis by Man-Whitney U-test. EPA: eicosapentaenoic acid; DHA: docosahexaenoic acid; AA: arachidonic acid; total n-3 PUFAs (Sum 20:5, 22:5, 22:3); total n-6 PUFAs (Sum 18:2, 20:3, 20:4, 22:3, 22:4).
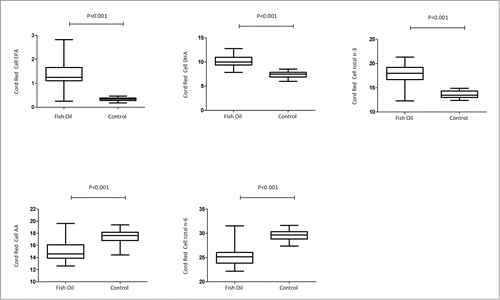
Figure 2. Regression analysis of CpG sites that co-associate with cord red cell total n-3 fatty acid levels. (A) qq plot of P-values from the association test between CpGs and total n-3 fatty acids did not deviate from the null hypothesis. (B) Scatterplots of the top 4 CpGs ranked by P-value for the association with total n-3 fatty acids suggested a dose-response relationship. (C) Histogram of the coefficients from the regression model indicated a small effect size.
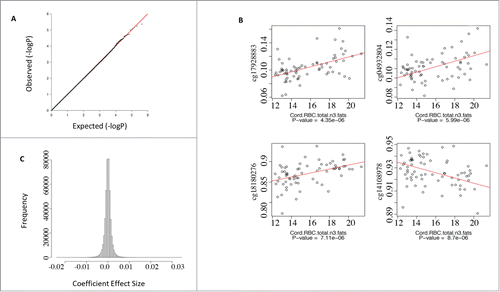
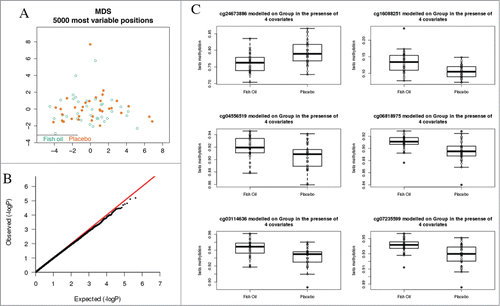
Figure 3. Comparative analysis of DNA methylation profiles between treatment and intervention groups. (A) MDS scaling analysis did not discriminate treatment groups. Samples are indicated by open circles and projected along axes of the first 2 principal components of the top 5000 most variable CpG sites. (B) qq plot of P-values from the moderated t-test between intervention and control groups did not deviate from the null hypothesis. (C) Boxplots of data from 6 top-ranked CpG sites for the between-group comparison did not indicate substantive differences in methylation levels. Boxplots show median and range.