Figures & data
Figure 1. Rice JAZ factors. (A) Jas domains of rice JAZ proteins. Amino acid sequences of the Jas domains were aligned using Clustal Omega (https://www.ebi.ac.uk/Tools/msa/clustalo/). The consensus sequence is indicated at the bottom. The 2 conserved positively charged amino acids shaded with yellow colors were substituted with alanine in the mutated JAZs. The other highly conserved amino acids are shaded in light blue. Names of JAZs are as reported in ref. 30 JAZ1, Os04g0653000 (OsTIFY3); JAZ2, Os07g0153000 (OsTIFY5); JAZ3, Os08g0428400 (OsTIFY6a); JAZ4, Os09g0401300 (OsTIFY6b), JAZ5, Os04g0395800 (OsTIFY9); JAZ6, Os03g0402800 (OsTIFY10a); JAZ7, Os07g0615200 (OsTIFY10b); JAZ8, Os09g0439200 (OsTIFY10c); JAZ9, Os03g0180800 (OsTIFY11a); JAZ10, Os03g0181100 (OsTIFY11b); JAZ11, Os03g0180900 (OsTIFY11c); JAZ12, Os10g0392400 (OsTIFY11d); JAZ13, Os10g0391400 (OsTIFY11e), JAZ14, Os10g0391801 (OsTIFY11f); JAZ15, Os03g0396500 (OsTIFY11g). (B) Expression of JAZ genes in rice tissues. The levels of respective JAZs were determined by RT-PCR. Arrows indicate bands specific to respective JAZs. RNA was extracted from root and shoot tissues of 1-week-old seedlings and basal shoot tissues (1 cm long) of 2-week-old seedlings grown on the MS-based medium.15 Reverse transcription was performed using the Quanti-Tect Rev transcription kit (Qiagen). First-strand cDNA was amplified by PCR using KOD-FX Neo (Toyobo) or EX-Taq (Takara). For amplification and subcloning of cDNA into pENTR-/D-Topo (Invitrogen), primers for JAZ1 (5′-caccATGGATCTGTTGGAGAAGAAG-3′ and 5′-TTACTGGGCCTTGCCCTCAG-3′), JAZ2 (5′-caccATGGCGGAGGAGCGGAGGAGAGACGAC-3′ and 5′-CTATCGCCGGGCGTACAGCGGCGCGG-3′), JAZ3 (5′-caccATGGAGAGGGATTTTCTTGGC-3′ and 5′-TCATATCTGTAACTTTGTGCTGGGGGGC-3′), JAZ4 (5′-caccATGGAGAGGGACTTCCTGGG-3′ and 5′-TCAGATTTGTAGCTTTGTACTGGGGGACTCC-3′), JAZ5 (5′-caccATGTCGACGAGGGCGCC-3′ and 5′-CTAGGACGCCGTGTGCTCCTCTTCCTTC-3′), JAZ6 (5′-caccATGGCTTCCGCGAAATC-3′ and 5′-TCATTGGCTCGATTCCTGCCG-3′), JAZ7 (5′-caccATGGCGGCTTCCGCGAGG-3′ and 5′-TCATTGGCCGCGTTCTATGGGCTTCACG-3′), JAZ8 (5′-caccATGGCCGGCCGTGC-3′ and 5′-TCATATCTCCTGCTTTATTGTCATCTCTTGGCC-3′), JAZ9 (5′-caccATGGCGTCGACGGATCCCAT-3′ and 5′-TCAGCGCGAGTGCATGTGTCCAA-3′), JAZ10 (5′-caccATGGCGATGGAGGGGAAGAGC-3′ and 5′-TCACAGCGCGATGGTGAGGCTGTC-3′), JAZ11 (5′-caccATGGCCGGTAGTAGCGAGCA-3′ and 5′-TCACAGGCTGAGAGTGGGGTTCACCT-3′), JAZ12 (5′-caccATGGCCGCCGCCGGCAGCAGC-3′ and 5′-TCAGAGCCCGAGCCATGTCGCCGGCTCA-3′) and for JAZ15 (5′-caccATGGACGCCGTCGGCGCAG-3′ and 5′-TCACTTTTGCTTCCTCTTTTGGAGGAATCG-3′), respectively, were used. After subcloning, mutations were introduced by site-directed mutagenesis, using primers for mJAZ3 (5′-GCCTCAAGCTGCAGCAGCATCTCTCG-3′ and 5′-CGAGAGATGCTGCTGCAGCTTGAGGC-3′), mJAZ4 (5′-CCCTCAAGCGGCGGCGGCATCCCTTG-3′ and 5′-CAAGGGATGCCGCCGCCGCTTGAGGG-3′), mJAZ6 (5′-GCCTCAGGCTGCGGCGGCGTCGCTTC-3′ and 5′-GAAGCGACGCCGCCGCAGCCTGAGGC-3′), mJAZ7 (5′-GCCTATTGCTGCGGCGGCATCACTCC-3′ and 5′-GGAGTGATGCCGCCGCAGCAATAGGC-3′) and mJAZ11 (5′-GCCCATCGCGGCGGCGGTGTCGCTGC-3′ and 5′-GCAGCGACACCGCCGCCGCGATGGGC-3′), respectively.
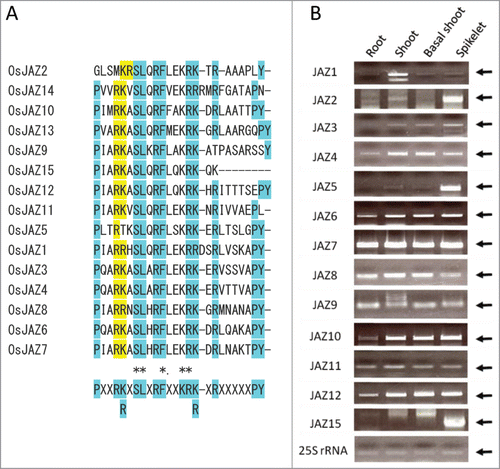
Table 1. Summary of aberrant morphogenesis observed in the transgenic lines
Figure 2. Morphology of spikelets in transgenic rice lines overexpressing mJAZ (A, B) A line carrying empty vector. (C, D) mJAZ3ox, line Y10. Note that an ectopic spikelet-like structure (E-Sp) has formed in a spikelet. To indicate internal tissues, outer and floral organs are separated in (B, D). (E) mJAZ6ox, Line1. Leafy glume-like organs (L-Gl) that exhibited curved structure along the apical-basal axis, resembling leaf blades, generated repetitively. (F) mJAZ7ox, Line5. (Inset) Internal tissues visualised by detaching glumes. No stamens or pistil have been formed. A-Gl, aberrant glume-like structure; E-Lo, elongated lodicule; E-Sp, ectopic spikelet-like organ; E-SL, elongated sterile lemma; L-Gl; leafy glume-like organ; Le, lemma; Le-l, lemma-like organ; Lo, lodicule; Pa, palea; Pe-l; pedicel-like organ; Pi, pistil; RG, rudimentary glume; SL, sterile lemma; St, stamen. For overexpression of mJAZ, respective cDNAs carrying the mutations were introduced into the binary vector pSMAHdN637L-GateA and used for Agrobacterium tumefaciens-mediated transformation of rice (Oryza sativa, cv Nipponbare) as described in ref. 24 Insertion of respective transgenes in the genome DNA was confirmed by PCR amplification. For preparation of an empty vector control, 1.7-kb DNA fragment containing ccdB and sequences for site-specific recombination (attR1 and attR2) were eliminated from the vector plasmid by digestion with XbaI and KpnI, followed by blunting and ligation.

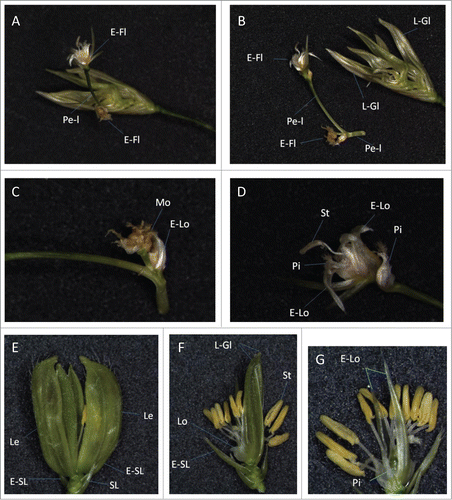
Figure 3. Repetitious formation of floral and outer organs in a spikelet of a transgenic rice. (A–D) A spikelet of a transgenic line (mJAZ11ox, Line A1). (A) A pedicel-like organ (Pe-I), carrying ectopic floral organs, formed at the center of the spikelet. (B) The pedicel-like organ was dissected from the center of the spikelet. Note that leafy glume-like organs (L-Gl) developed repetitiously. (C) A magnified view of the ectopic floral organ formation at the base of the pedicel-like organ. Mosaic of pistil- and lodicule-like organs (Mo) formed on an axillary branch. (D) A magnified view of the ectopic floral organs formed on the apex of the pedicel-like organ. Note the position of the pistils inside and outside of the repetitively formed elongated lodicules (E-Lo). (E–G) A spikelet of a transgenic line (mJAZ11ox, Line 4). (E) Repetitious formation of glumes in the spikelet. (F, G) Internal organs of the spikelet, visualised by removing 2 lemmas (F) and leafy glume-like organs (G; magnified). E-Fl, ectopic floret; E-Lo, elongated lodicule; E-SL, elongated sterile lemma; Le, lemma; L-Gl; leafy glume-like organ; Lo, lodicule; Pe-l; pedicel-like organ; Mo, mosaic organ; Pi, pistil; SL, sterile lemma; St, stamen.