Figures & data
Figure 1. Showing a comparison of volatiles production from control (black) and heat stressed (red). Peaks 1, 2, 3 are isoprene, 2-methyl butanenitrile and β-Ocimene respectively as compared with >90% similarity match to NIST/EPA/NIH Mass Spectral Library, Data version: NIST 11, Software version 2.0. Above peaks were confirmed with 3 biological samples.
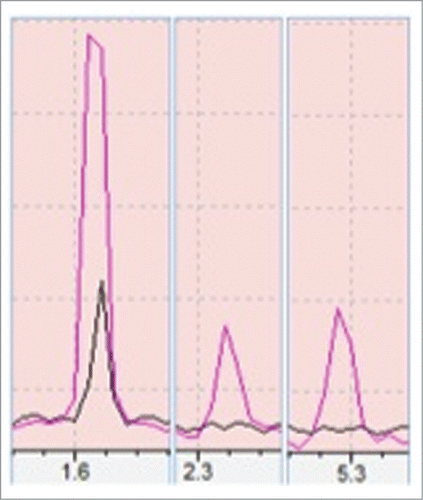
Figure 2. Showing comparison of peaks produced by total lipid extracted from control (black) and heat stressed plant (red) through GC-MS. Only the peaks having >90% similarity with the GC-MS NIST/EPA/NIH Mass Spectral Library, Data version: NIST 11, Software version 2.0 were shown here. In the above figure each part is with different magnification to clearly show the differences. The peaks 1 to 16 are phenol, α- Cubebene, Copaene, β-Elemene, α-Gurjunene, Caryophyllene, α-Guaiene, Humulene, Germacrene D, α-Selinene, gamma-Elemene, Cadina-1(10),4-diene, Germacrene-D-4-ol, Phytol acetate, Phytol and Octacosane. The differences in peaks were inferred with triplicate of technical replications.
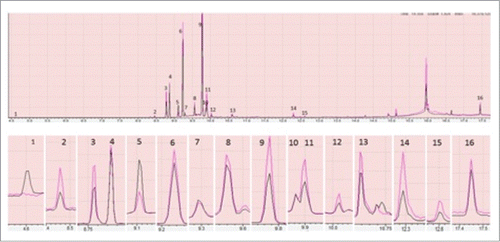
Table 1. Volatile compounds produced by heat stressed (HS) Hymenaea plants
Table 2. Lipid analysis of heat stressed (HS) Hymenaea plant as identified by GC MS
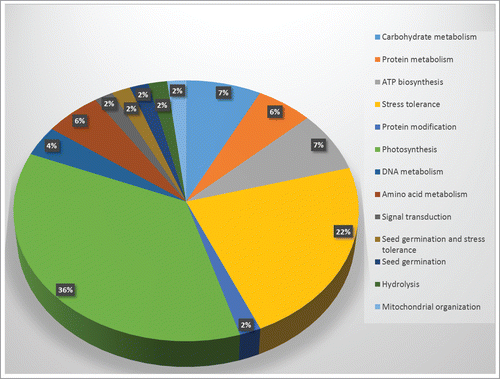
Table 3. Differentially expressed proteins in Hymenaea plants after heat stress as identified by 2-D gel electrophoresis
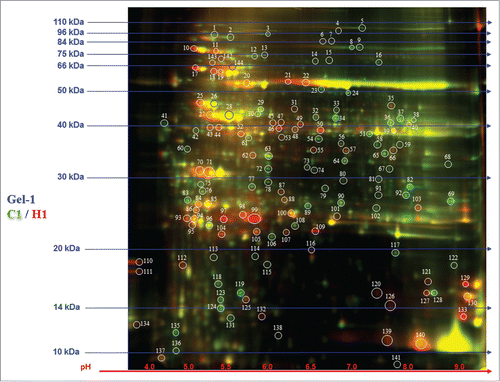
Figure 3. 2D-DIGE analysis of Hymenaea leaf proteome. Protein samples of Hymenaea control plants and heat stressed plants were differentially labeled with Cy3 (green) and Cy5 (red) respectively. After mixing the 2 labeled proteins in equal ratios, they were first subjected to isoelectric focusing on a IPG strip, pH 3–10, and then on a 12.5% SDS-PAGE. The isoelectric point (pI) and molecular mass (kDa) are marked. Color coding: green spots indicates that the protein abundance is high in Cy3 (control), red spot indicates that the protein abundance is high in Cy5 (heat stressed) samples; yellow spots indicates that the protein abundance is similar in both the cases. Protein identifications of selected spots are shown in .