Figures & data
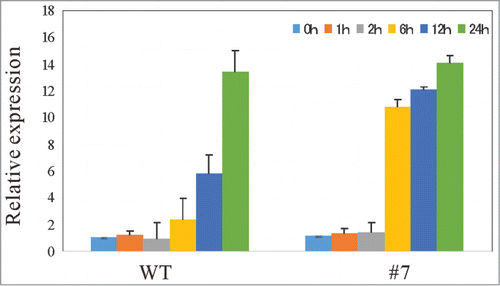
Figure 1. The domain structures of Arabidopsis VAP proteins. Domain structures of Arabidopsis VAP (PVA) proteins were predicted with the SMART program (http://smart.embl-heidelberg.de/). On the basis of the molecular structures and similarities of VAP proteins, we categorized 10 Arabidopsis VAP proteins into 5 subgroups. The abbreviations of domains used in this figure are as follows: CCD: coiled-coil domain, MSP: major sperm protein domain, TIR: Toll/interleukin-1 receptor homology domain TM: transmembrane domain. The numbers indicate the length of molecules in amino acid.
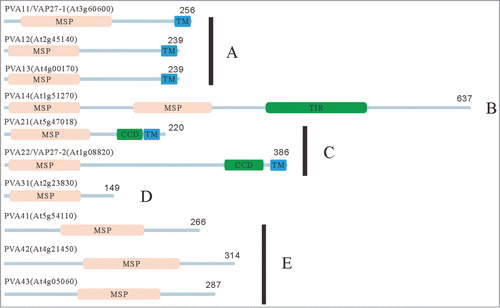
Figure 2. Localization of GFP-PVA31 in Arabidopsis. The confocal images of GFP fluorescence (A, C) and merged DIC and GFP-fluorescence (B, D) were captured in epidermal cells of hypocotyls (A, B) and mesophyll cells of cotyledon (C, D) of Arabidopsis plants transiently expressing GFP-PVA31. The images of GFP fluorescence in epidermal cells (E). The confocal images of GFP-PVA31 (F), mRFP-VAMP722 (G) and merged image of F and G (H) in epidermal cells of hypocotyl. Arrows indicate the mobile punctate structures of GFP-PVA31. Bars = 50 μm (A) , 20 μm (C) and 10 μm (H, K).
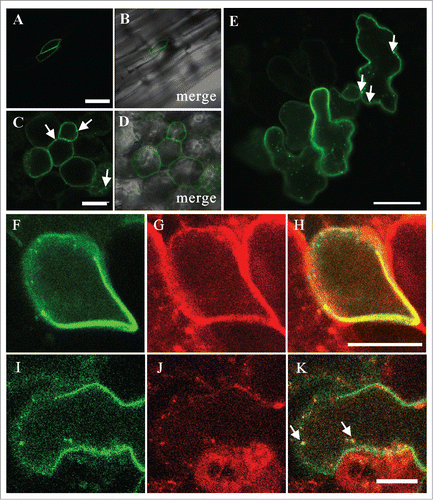
Figure 3. Interaction of PVA31 and VAMP proteins. Yeast two-hybrid assay were performed to confirm between PVA31 and Arabidopsis VAMP proteins. PVA31 and VAMP/711/721/722/724/727 were fused with activation domain (AD) and DNA binding domain (BD) of the transcription factor GAL4, respectively. The yeast strains co-transformed with PVA31 and indicated VAMPs plasmids were grown on selective media lacking leucine, tryptophan and histidine and adding 3-AT. Positive control: pGADT7-T-antigen and pGBKT7-p53. Negative control: pGADT7-T-antigen and pGBKT7-Lamin C.
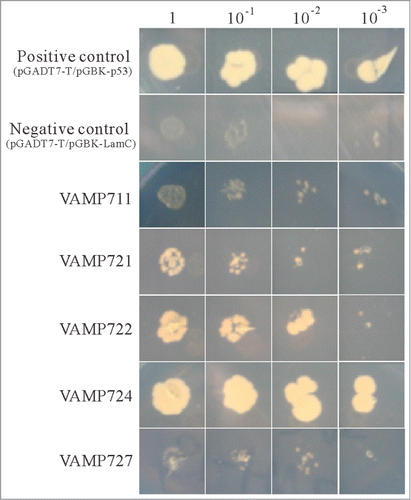
Figure 4. Age-dependent senescence symptoms in PVA31-overexpressing plants. A quantitative RT-PCR analysis of SAG12 transcripts isolated green rosette leaves of WT and PVA31-overexpressing plants growing for 4-weeks. UBQ10 was used as an internal control. Each vertical bars represented the mean ± standard error (3 leaves from each plant were used for the analysis) (A). Senescence phenotype of the rosette leaves of WT and PVA31-overexpressing plants (lines #7 and #13) grown for 8 weeks. The whole rosette leaves of 8-weeks-old plants were detached and aligned in leaf age order (B). The senescence symptom of rosette leaves. Arrowheads indicate the senescence symptom in representative leaves (C). The percentages of senescent leaves of each plant (WT or PVA31-overexpressing lines #7 and #13) with respect to total leaf numbers are presented. Error bars indicate standard deviations. Ten plants from each line were evaluated according to the senescence symptom shown in C (D). The chlorophyll content was measured before and after dark treatment in detached leaves. The difference between WT and PVA31-overexpressing line was indicated by an asterisk (P < 0.01) (E).
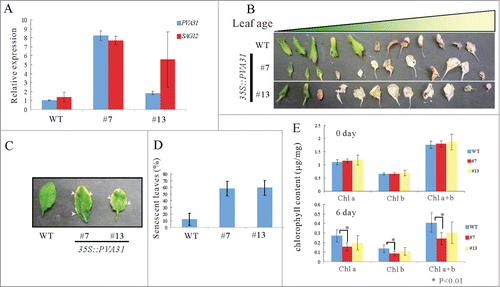
Figure 5. Expression of marker genes for SA-mediated biotic stress responsive pathway are upregulated in PVA31-overexpressing plants with elicitor application A quantitative RT-PCR analysis of PR-1 gene was performed with total RNA isolated from a whole 14-day-old WT plant at various time points (0, 1, 6, 12, and 24 h) following flg22 treatment. UBQ10 was used as an internal control. Each vertical bars represented the mean ± standard error (3 leaves from each plant were used for the analysis).