Figures & data
Figure 1. Chromosome 13 painting combined with FISH of the pluripotency marker gene GOF. (A1) Wide-field, digital, fluorescence microscopy from a bovine fetal fibroblast metaphase spread after 3-color FISH shows DAPI stained chromosomes (blue) painted chromosomes 13 (red), the pluripotency marker gene GOF (green) and α-satellite DNA clusters in pericentromeric heterochromatin (white). (A2) Magnification of the boxed carrier chromosome 13 demonstrates the integration of GOF on 13q near the pericentromeric heterochromatin.
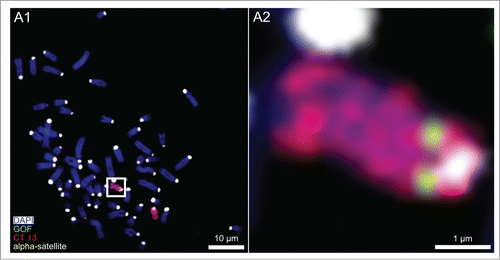
Figure 2. Variability of higher order chromatin arrangements of the pluripotency marker gene GOF, its carrier chromosome territory (CT) 13 and the non-carrier homolog. Panels A-F. CSLM images were recorded from 6 nuclei A-F representative for the variability of radial nuclear arrangements of GOF (green), its carrier and non-carrier chromosome territories 13 (red): 2 fibroblast nuclei (A and B), 2 embryonic nuclei from cloned embryos at day 2 (C and D), and 2 nuclei from cloned embryos at day 4 (E and F). DAPI stained DNA is presented in gray. X/Y-sections (A1-F1) and X/Z sections (A2-F2) were taken at positions including the painted carrier chromosome territory with the GOF signal. Two white arrows in A1-F1 indicate the site, where the X/Z section was taken perpendicular to the X/Y section. Correspondingly, 2 white arrows in A2-F2 indicate the site, where the X/Y section was taken perpendicular to the X/Z section. A3-F3 show partial 3D reconstructions of the same nuclei presenting the location of the carrier and non-carrier chromosome territories 13. A4-F4 show enlarged, virtual sections of the carrier chromosome territory with GOF signals representing the nearest position of GOF to the DAPI stained nuclear border. Bars: 3 μm for A1-F1, A2-F2, A3-F3; 2 μm for A4-F4.
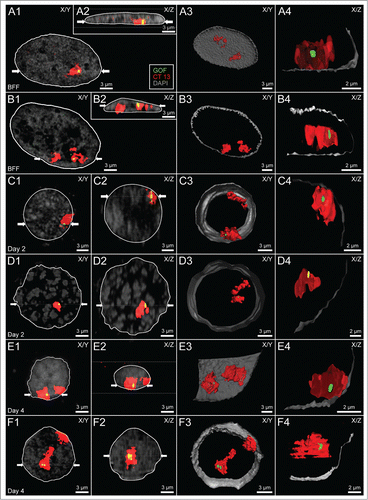
Table 1. Chromosomal gains and losses in cloned and IVF bovine embryos
Figure 3. (A) Volume measurements of bovine fetal fibroblast (BFF) nuclei, day 2 and day 4 embryonic nuclei. Compared with the volumes of fibroblast nuclei, a severalfold increase was noted for day 2 nuclei, whereas a decrease of nuclear volumes back to the level of fibroblast nuclei was noted in day 4 embryos. (B and C) Flat-ellipsoidal shaped fibroblast nuclei adopt a roundish shape in cloned embryos. (B) Scheme for the determination of nuclear roundness factors and relative radial distances. A roundness factor (RF) = 0 represents an extremely flat nucleus with zero axial extension, RF = 1 a perfect round shape. (C) Compared with RFs of fibroblast nuclei, RFs of nuclei in day 2 and day 4 embryos showed a severalfold increased roundness factor.
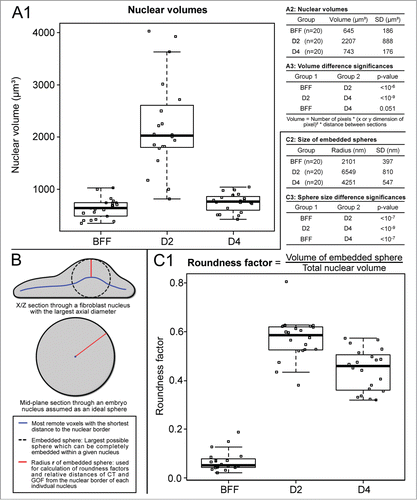
Table 2. Statistical analysis of differences between absolute, mean distances of GOF and chromosome territory (CT) 13 gravity centers to the nuclear border in bovine fetal fibroblasts (BFFs), day 2 and day 4 cloned embryos
Table 3. Statistical analysis of differences between relative, radial positions of GOF and chromosome territory (CT) 13 gravity centers in bovine fetal fibroblasts (BFFs), day 2 and day 4 cloned embryos
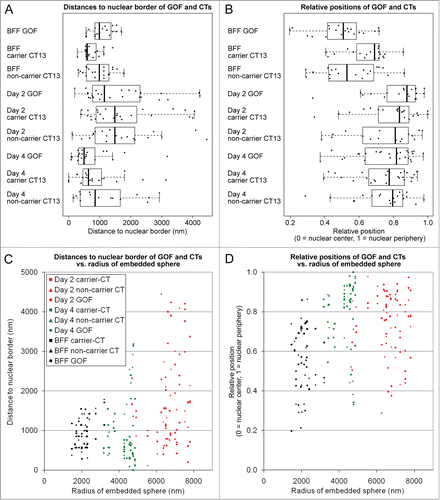
Figure 4. Quantitative analysis of radial nuclear arrangements of the pluripotency reporter gene GOF, its carrier chromosome territory (CT) 13 and the non-carrier homolog. (A) Absolute 3D distances from the 3D reconstructed nuclear border. (B) Relative 3D positions between the center of the nucleus (0) and the 3D reconstructed nuclear border (1). Combined box/scatter plots of GOF, the carrier and non-carrier chromosome territories CT 13 in fibroblast nuclei, day 2 embryonic nuclei and day 4 embryonic nuclei observed in cloned embryos at the onset of major embryonic genome activation and shortly thereafter demonstrate an extensive internuclear variability of these measurements. (C and D) present the data set shown in (A and B), respectively, in relation to the largest sphere, which could be embedded in each given nucleus (compare and ). Note that the variability of absolute distance measurements increased with the size of the embeddable sphere (C), whereas a similar variability was detected with respect to the relative radial positions (D).