Figures & data
Figure 1. Developmental Analysis of Symplekin Mutants (A). Schematic of Symplekin locus, gene structure and mutations. Symplekin is located on the 3rd chromosome in close proximity to the essential genes madm and cas. Features of the Sym gene structure are represented by intensity of grey: intergenic (light), UTRs (medium) and ORF (dark). Transcription start sites are represented with an arrow above the gene and triangles below indicate the transposon insertion sites. Outlined boxes depict the nature of each Sym mutant. The genomic deletion in Sym152 is shown above the locus diagram. The EY P element inserted in SymEY and the internal deletion of the EY P element leaving 206 nts in the Sym 5'UTR resulting in Sym58 are represented below the Sym gene structure diagram. (B). Visual representation of Sym mutant viability phenotypes. Circles indicate the proportion of Symallele mutant (black) and control heterozygous sibling (grey) flies obtained in each experiment. The expected number of control siblings is 2/3. Thus, 1/3 black indicates full viability and the absence of black indicates lethality. The percentage of observed flies within the mutant class is labeled inside each circle. Note that the observed precise excision (Sym53) class exceeded the expected ratio as a wild type fly is healthier than control siblings carried over a balancer chromosome. For the EY genotypes, (p) indicates a paternal and (m) indicates a maternal contribution of the transposon with the other parent providing the Df chromosome. For all of the other genotypes, the Df chromosome was maternally provided. (C). Visual representation of the Sym58 delayed eclosion phenotype. The line graph represents the daily average number of mutant and control sibling flies observed within one generation. (D). Symplekin protein levels in Symplekin mutant embryos. Equal amounts of total protein from 16-20 hour embryos were resolved by SDS gel electrophoresis and Symplekin detected by Western blotting. The mutant Sym152 (lane 2) does not express zygotic symplekin and indicates the amount of maternal Symplekin remaining at this stage. The * indicates a cross-reacting band. (E). Extracts from 3rd instar larvae of each genotype were analyzed by Western blotting. 3-fold dilutions of each genotype were analyzed for Symplekin (top) and the blot re-probed for tubulin (bottom).
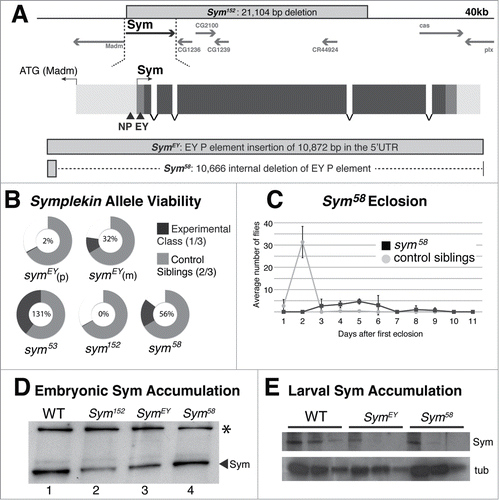
Figure 2. Depletion of Symplekin protein results in misprocessed RNA (A). S1 nuclease protection assay analysis of histone mRNA species in Symplekin mutants. The diagram indicates the location of the H2a probe. Total RNA extracted from wandering 3rd instar larvae or adult ovaries was hybridized with the 3'-labeled (star) DNA probe (P) and then incubated with S1 nuclease, which digests single stranded nucleic acids. The probe is complementary to the H2a mRNA, H2a-H4 intergenic region up to the H4 HDE and also contains vector sequence (3' grey region). Properly processed histone H2a mRNA is cleaved between the stem loop (SL) and histone downstream element (HDE), protecting 340 bp of the DNA probe from S1 digestion (W). The H2a gene contains multiple cryptic poly(A) signals that are utilized in animals with disrupted histone pre-mRNA processing. These longer mis-processed polyadenylated mRNAs protect several discrete species (M). Any transcripts extending beyond the region of complementarity protect a single read-through product (R) that is distinct from undigested probe (P) and whose 3' end(s) are likely heterogeneous. Protected probe products were resolved on an acrylamide urea gel and visualized by autoradiography. (B). Processing of polyadenylated mRNAs in Sym mutants was assessed by RT-PCR with primers flanking the canonical cleavage site for three mRNAs, rp49, sop and pgk. The black bar above each gene diagram represents the amplified PCR product. Black boxes represent exons, the grey box represents the 3'UTR and the dashed line represents the downstream intergenic region, which accumulates in the Symplekin mutant. pgk transcripts were detected in each preparation. Note that the amplified product in the pgk gene spans an intron that is not present in the cDNA template but is in the genomic DNA PCR control.
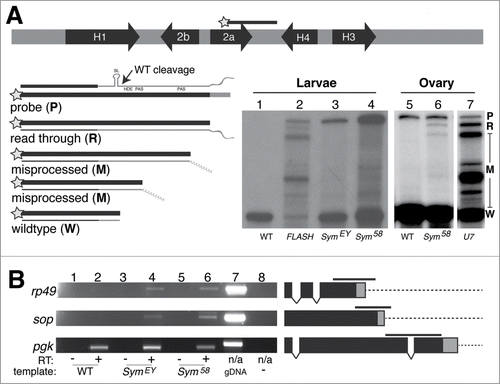
Figure 3. Symplekin concentration in the HLB is dynamic during development. (A and B). Projected confocal images of Symplekin localization in a syncytial embryo (A) and salivary gland nuclei (B). The presence of Symplekin in the HLB was determined by co-localization with the constitutive component Mute. Cyclin E/Cdk2 activity was monitored by staining with MPM-2, an antibody that recognizes phosphorylated epitopes in the HLB. Note that in the embryo, Sym enrichment was detected in both rapidly cycling somatic (yellow arrow) cells and germline (white arrow) cells that are not cycling. Both types of cells have MPM-2 positive HLBs whereas in the salivary gland nucleus, neither Sym nor MPM-2 signal was present in the large Mute positive HLB (red arrowhead). Scale bars = 10 microns. The inset shows a higher magnification image of a somatic cell. (C). Quantification of Sym HLB enrichment in endocycling stage 8 ovarian follicle cells. Ovaries were stained for Mute (blue), Sym (green) and MPM-2 (red). HLBs were defined by Mute staining, and we assessed the HLBs for co-localization with Sym and/or MPM-2. A representative projected image shows each class: Mute only (blue arrow), Mute + MPM-2 (red arrow), Mute + Symplekin (green arrow) and Mute + MPM-2 + Symplekin (yellow arrow). Quantification of individual ovaries were averaged and presented as a Venn diagram (n=6, average number of cells scored per ovary was 93 +/- 21.7). The errors represent the standard deviation.
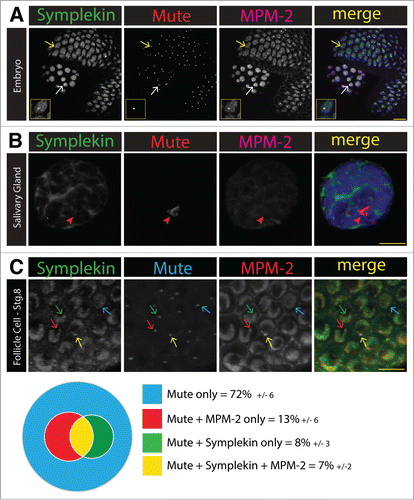
Figure 4 (See previous page). Symplekin localizes to the tri-cellular junction in stage 10B ovary follicle cells. (A–D). Projected confocal images of the localization of Symplekin in stage 10B ovarian follicle at the tricellular junctions. (A) A V5-tagged Lsm11 replacement strain that has a wild-type phenoptype.Citation37 Note that these cells are stained with anti-Dlg and anti-V5 antibodies in the same channel to simultaneously identify cell boundaries (yellow arrow) and V5-Lsm11 in the HLB (pink arrow). (B) Visualization of Symplekin and Discs large in wild type stage 10B ovarian follicle cells by structured illumination fluorescence microscopy. Note that Symplekin accumulates at the cell cortex where three cells come together (yellow arrow). (C) Follicle cells expressing HA tagged-Sym stained with anti-HA antibodies. (D) Wild type follicle cells stained with anti-Sym and anti-Gliotactin, a tricellular junction marker. (E–F). Localization of additional RNA processing factors in stage 10B ovarian follicle cells. Wild type follicle cells stained with anti-CPSF73 and anti-Dlg antibodies (E) or anti-CstF64 and anti-Dlg (F). Note that components of the cytoplasmic polyadenylation complex (Sym and CPSF-73) localize to the tri-cellular junction (yellow arrow), whereas CstF-64 does not (white arrow).(G–H). Projected confocal images representing the relationship between Sym and RNA binding protein yps in stage 10B ovarian follicle cells. Wild-type cells were stained with Yps and Dlg (G). Yps is enriched at the tricellular junctions (yellow arrow). ypsJM2/Df(3L)BK9 mutant cells were stained for Sym and Dlg (H). In the yps mutant Sym is diffuse in the cytoplasm. Yellow bars = 10 microns. Arrow indicates magnified cell shown in inset in panels C-H (yellow box) (I). Alignment (ClustalW2) of the amino sequences of human ZONAB and Drosophila yps.
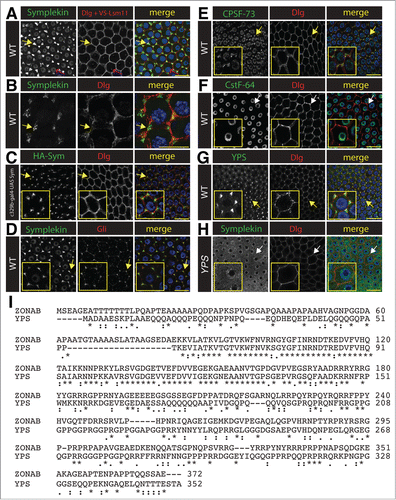
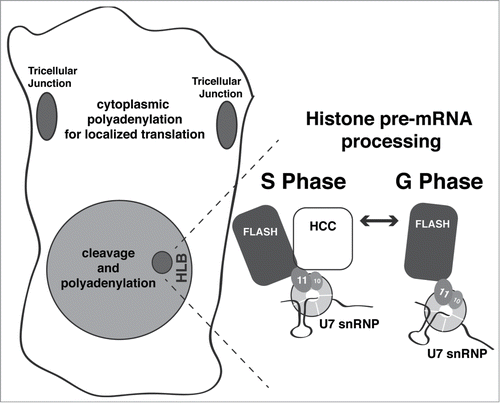
Figure 5. Summary of Symplekin subcellular localization and model of HLB dynamics. Symplekin localization observed in this study is summarized in the cell drawn on the left. Grey shading represents the diffuse Symplekin accumulation in the nucleus, consistent with its role in cleavage-polyadenylation. The darker grey indicates sites of Symplekin concentration: in the tricellular junctions, where it likely participates in cytoplasmic polyadenylation leading to localized translation; and in the HLB, where it is required for histone mRNA biosynthesis. The expanded HLB depicts fluctuating concentration of Sym during the cell cycle. During S phase or when there is high activity of Cyclin E/Cdk2, Symplekin (and likely the HCC) localizes to the HLB and interacts with a unique surface generated by an interaction between FLASH and the U7 snRNP component Lsm11, creating the active form of U7 snRNP.