Figures & data
Table 1. Bioassay results for the phenol/chloroform group: first and second experiments
Figure 1. Nucleic acid analysis and formation of recPrP nanostructures. (A) Electropherogram of inoculum 8 (DNase I-digested inoculum 2) analyzed using a small RNA assay chip, showing 2 distinct populations of RNA with average lengths of approximately 27 and 55 nucleotides. Approximate quantities of oligonucleotides measured in each peak are 0.5 ng for the marker, 0.2 ng for the first peak (∼27 nt) and 0.07 ng for the second peak (∼55 nt). (B) Electropherogram of DNase I-digested inoculum 2 analyzed using a large RNA integrity assay chip, showing the absence of large cellular RNA and degradation products. (C) Inoculum 2 analyzed using a small RNA assay chip, showing the presence of cellular DNA (sizes ≤ 300 nucleotides). Approximately 37 ng of DNA is observed for 0.16 mg brain equivalent loaded. (D) Electron micrograph of inoculum 8 after negative staining, indicating the presence of ovoid and spherical aggregates of PrP with diameters of 7-40 nm. (E) Electron micrograph of control inoculum 7 (Ph/Ch + α-recPrP monomers), showing the absence of PrP aggregates (scale bar, 100 nm). For (A-C) 1 μl of sample was loaded onto the chip, which is approximately equal to 8 ng equivalent PrPSc and approximately 0.16 mg brain equivalent.
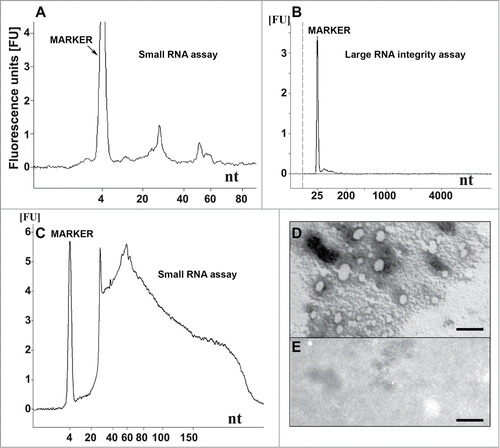
Figure 2. Nucleic acid analysis from the second reconstitution experiment. Electropherogram of inoculum D, showing the same distribution of small RNA populations as in inoculum 8. Approximate quantities of each peak are 0.5 ng for the marker, 0.18 ng for the first peak and 0.05 ng for the second peak. A 1 μl sample was loaded onto the chip, which is approximately equal to 8 ng equivalent PrPSc and approximately 0.16 mg brain equivalent.
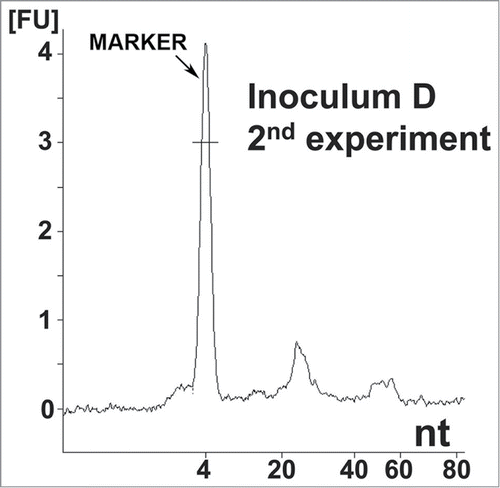
Table 2. Bioassay results for the guanidinium-HCl (GndHCl) group
Figure 3. Testing for PrPSc seeding activity in phenol/chloroform extracts and bioassay samples (A) Testing phenol/chloroform extracts (Ph/Ch) from a crude preparation of 263K scrapie-associated fibrils that had been purified using a previously published protocolCitation31 for residual PrPSc seeding activity using serial PMCA (the third and fourth PMCA rounds are shown). Lanes 1: PK digested 263K scrapie hamster brain homogenate containing 5 × 10−7 g of 263K hamster brain tissue. Lanes 2–8: Reference PMCA samples seeded with 1 × 10−12 (lanes 2 and 3), 1 × 10−11 (lanes 4 and 5), 1 × 10−10 (lanes 6 and 7) or 1 × 10−9 g 263K hamster brain tissue (lane 8). Lanes 9 and 10: Phenol/chloroform extracts isolated from uninfected hamster brain. Lanes 11 and 12: Ph/Ch extracts isolated from 263K scrapie hamster brain (corresponding to ∼500 ng 'equivalent PrPSc' per lane, i.e., to an approximately 2.5-fold amount of the intracerebral inoculum administered to hamsters, which was 200 ng 'equivalent PrPSc'). Lanes 13 and 14: Ph/Ch extract isolated from 263K scrapie hamster brain spiked with 1 × 10−11 g of 263K scrapie brain tissue following phenol/chloroform extraction. (B) Testing various bioassay inocula for the amount of residual or newly produced PrP seeding activity using serial PMCA. Lanes 1, 8, 15 and 19: PK-digested 263K scrapie hamster brain homogenate containing 5 × 10−7 g of 263K hamster brain tissue. Lanes 2–7: Reference PMCA samples seeded with 1 × 10−12 (lanes 2 and 3), 1 × 10−11 (lanes 4 and 5), 1 × 10−10 (lane 6) or 1 × 10−9 g 263K hamster brain tissue (lane 7). Lanes 9–14: Negative controls without seeding material (i.e., PMCA with normal hamster brain homogenate only). Lane 16: PMCA seeded with 10 μl of inoculum 6 (Ph/Ch + β-rec PrP oligomers). Lane 17: PMCA seeded with 6 μl of inoculum 7 (Ph/Ch + α-rec PrP monomers). Lane 18: PMCA seeded with 6 μl of inoculum 8 (Ph/Ch + DNase I + β-rec PrP oligomers). Lanes 20–22: Same samples as in lanes 16–18, spiked with 1 × 10−10 g of 263K scrapie brain tissue.
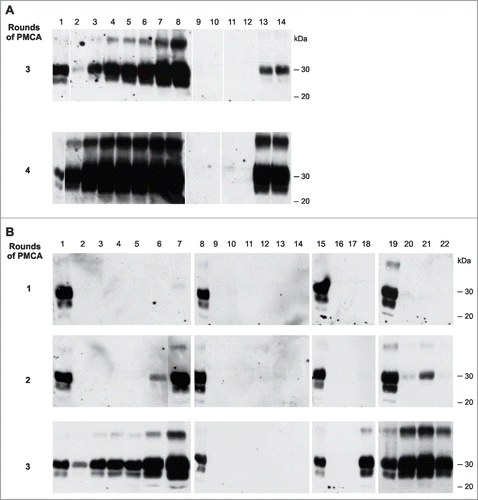
Figure 4. In-vitro amplification of PrPSc is fully reconstituted by the addition of synthetic polyA RNA. Serial PMCA (5 rounds) was performed with nuclease-treated PrPSc seeds purified from 263K scrapie hamster brains and differently pretreated normal hamster brain homogenates (NBHs) as substrates. NBHs were digested with (A) Benzonase, (B) Benzonase subsequently spiked with PolyA RNA (200 μg/ml), or (C) RNAse A. The amount of original seeding material corresponded to an extract from 1 × 10−6 g 263K scrapie hamster brain homogenate. After each PMCA round, samples were diluted 1:5 in the corresponding substrates. P: PK digested 263K scrapie hamster brain homogenate containing 5 × 10−7 g of 263K hamster brain tissue, which served as a western blot positive control.
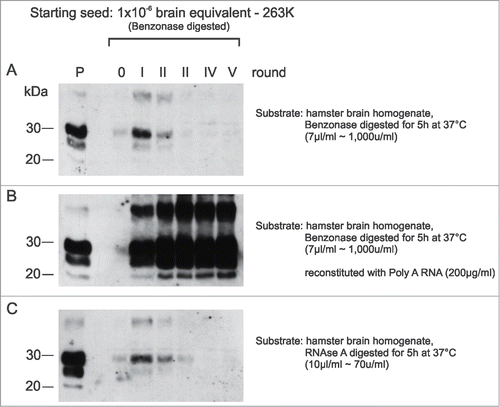
Figure 5. Histopathology and PrP-immunohistochemistry of brain sections from hamsters challenged with inoculum 8 (hamster 8.1: 151 dpi). Hematoxylin and eosin (H&E)-stained samples (2 columns on the left) and PrP immunohistochemistry using anti-PrP-antibody SAF32 (right column). (A-C) Control hamster, (D-F) hamster challenged with inoculum 8.1 (terminal phase, 76 dpi), (G-I) hamster challenged with SAFs of 263K scrapie agent (terminal phase, 76 dpi). The brain of the hamster challenged with inoculum 8.1 showed intense tissue damage of the hippocampus, extensive spongiosis, severe gliosis and neuronal loss (D, E). PrPSc immuno-detection (F) displayed a diffuse and widespread distribution that has not previously been observed in hamsters challenged with 263K SAFs (I).
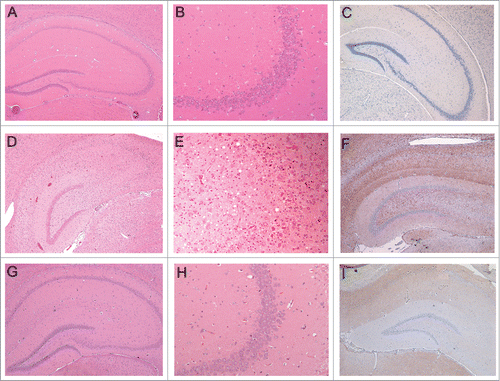
Figure 6. PrP-immunohistochemistry of hippocampus sections from infected hamsters using 3F4 and SAF32 anti-PrP antibodies. (A, B) 263K scrapie agent (IP: ∼80 dpi). (C, D) Inoculum 8.1 (1st passage; IP: 151 dpi). (E, F) Inoculum 8.1 (2nd passage; IP: 76 dpi). (G, H) Inoculum 8.2 (1st passage; IP: 246 dpi). I, J, Inoculum 8.2 (2nd passage; IP: 76 dpi). (K, L) Inoculum D (1st passage; IP: 148 dpi). The intensity of labeling for all passages was consistently stronger in the sections of animals infected with inocula 8.1, 8.2 and D compared with those of animals inoculated with 263K scrapie agent. A peculiar lamination pattern that had not previously been described in scrapie pathology can be observed in the molecular layer for inocula 8.1, 8.2 and D. This lamination is more clearly visible with the 3F4 antibody (arrows).
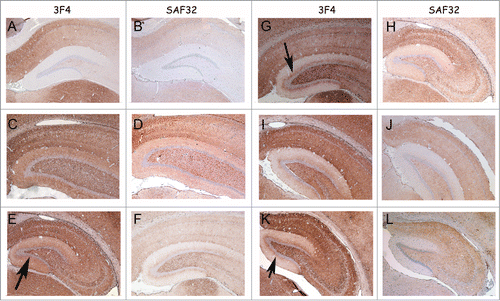
Figure 7. The lamination pattern observed in the hippocampal molecular layer corresponds to differential PrP labeling on synapses from specific areas. PrP-immunohistochemistry (3F4 antibody) of hippocampal sections from infected hamsters. (A, B) 263K scrapie agent (IP: ∼80 dpi). (C, D) inoculum 8.1 (3rd passage; IP: ∼80 dpi). (E, F) inoculum D (2nd passage; IP: ∼80 dpi). Lamination pattern corresponding to synaptic PrP immunostaining. (B, D, and F) Granular/punctate PrP depositions. HI (hilus), GC (granule cells), IZ (inner zone of the molecular layer), OZ (outer zone of the molecular layer). Magnification: (A) (x100), (B) (x400) (C and E) (x50), (D and F) (x200).
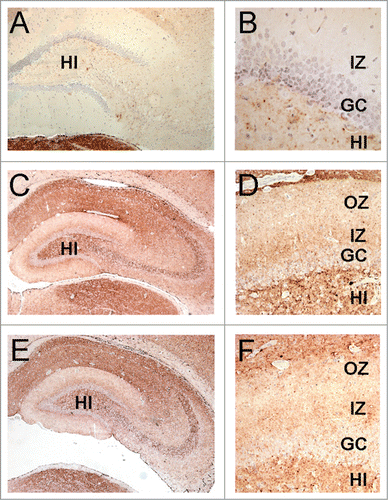
Figure 8. Detection of PrPSc in the brains of hamsters from the first and second passage (data from the first reconstitution experiment). (A) Brain PrPSc profile of infected animals from first reconstitution experiment. Inocula 9 (lane 3), 8.1 (lane 4; 151 dpi) and 8.2 (lane 5; 246 dpi). Lane 1: Positive control from 263K scrapie hamster; lane 2: control sample from healthy hamster. (B) Brain PrPSc profile of infected animals from first reconstitution experiment. Inocula 2* (lane 3; environmentally contaminated animal), 6 (lane 4), 7 (lane 5), 11 (lane 6) and 15 (lane 7). Positive control from 263K scrapie hamster; lane 2: control sample from healthy hamster. A selection of animals that died of causes not related to scrapie or deliberate infection are represented here. (C) First passage: WB profile of 2 distinct hamsters inoculated with BH of animal 8.1 (lanes 3 and 4); WB profile of 2 distinct hamsters inoculated with BH of animal 8.2 (lanes 5 and 6).

