Figures & data
Figure 1. Alignment of the RhoA, RhoB, RhoC, Rac1 and Cdc42 amino acid sequences. G motifs are marked with a blue box. Switch I, switch II, the insert region and the hypervariable C-terminus are highlighted in gray, sites for lipid modification are underlined. Sequence differences in RhoB and RhoC compared to RhoA are shown in red. Secondary structures (α-helices as cylinders, β-strands as arrows) are referring to RhoA and indicated below the amino acid sequence.
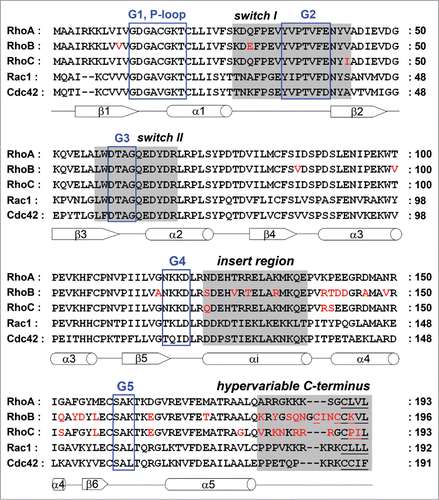
Figure 2. 3D structures of the RhoA, RhoB and RhoC G domain. (A–C) Ribbon representation of (A) RhoA (4–180 aa; Protein Data Bank (PDB) ID: 1FTN),Citation75 (B) RhoB (4–185 aa; PDB ID: 2FV8)Citation76 and (C) RhoC (3–179 aa; PDB ID: 2GCN).Citation77 Structures are shown in the same orientation. Switch I, switch II and the insert region are highlighted in gray, GDP is shown in yellow sticks. Residues marked in cyan differ among RhoA/B/C (underlined labeling in A) and are different compared to Rac1/Cdc42 (labeling in C). (D–F) Zooms of A–C highlighting that RhoA and RhoB contain at position 43 a valine, whereas RhoC has an isoleucine which has slightly bulkier side chain which underlies isoform-specific GEF- and effector binding (see text). (G–I) Electrostatic potential of the solvent accessible surface of the G domains of (G) RhoA, (H) RhoB and (I) RhoC represented in the same orientation as in A–C, lower panels show cognate structures rotated by 180°. Note that the hypervariable C-termini are not included in this representation. Electrostatic potential was calculated using the Adaptive Poisson-Boltzmann Solver (APBS) software, combined with the PDB2PQR server, at a pH of 7.5 and a threshold of ± 5 kTe−1 (red – negative charge; blue – positive charge).Citation78 Figures were prepared with PyMol (PyMol Molecular Graphics System, Schroedinger, LLC).
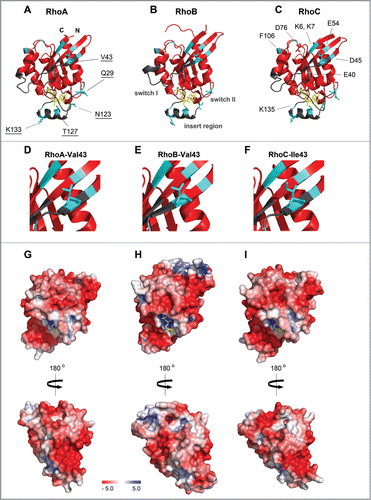
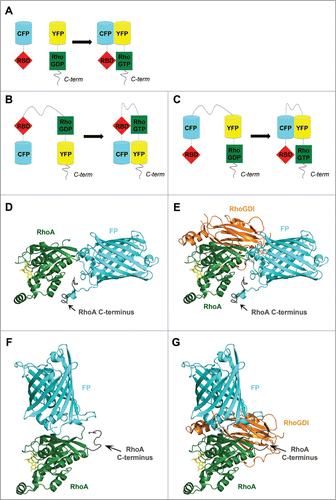
Figure 3. The structure of RhoGTPase FRET biosensors. Schematic representation of (A) an intermolecular FRET probe and (B and C) intramolecular FRET probes based on (B) Yoshizaki et al.Citation62 and (C) Pertz et al.Citation63 (D–G) The insertion of a fluorescent protein (FP) in the RhoA G domain may lead to mislocalization of the RhoA C-terminus and may impair binding of interaction partners such as RhoGDI. Homology model, calculated using the Phyre2 protein structure prediction server,Citation79 of RhoA-(1–180 aa)-FP-RhoA-C-terminus-(181–193 aa) alone (D) and in complex with RhoGDI (E) as well as of FP-RhoA-(1–193 aa) alone (F) and in complex with RhoGDI (G). Models are based on the structure of RhoA-GDP (PDB ID: 1FTN)Citation75 and GFP (PDB ID: 1H6R)Citation80 as top-scoring prediction events. Position of RhoGDI and GDP (from the complex Rac1-GDP-RhoGDI, PDB ID: 1HH4)Citation81 was obtained through the overlay of Rac1 and RhoA. GDP in yellow sticks; RhoA in green; FP in cyan, GDI in orange. Figures were prepared using PyMol.