Figures & data
Figure 1. Evolution of junctional architecture, and the molecular complexity of vertebrate junctions. Simplified schemes showing the organization of the apical junctional complexes of polarized epithelial cells in insects (as an example of invertebrates) and vertebrate organisms. The canonical functions (polarity, barrier, adhesion) of each type of junction (SAC = sub-apical complex/marginal zone, zonula adhaerens (ZA), septate junction, tight junction, desmosome) are indicated on the left of the respective junction. E-cadherin based junctions along the lateral contacts of epithelial cells (puncta adhaerentia) have a composition similar to that of punctate junctions between filopodial tips, e.g they contain a classical cadherin, and catenins (p120ctn, β-catenin, α-catenin), but not PLEKHA7 and afadin. On the right, a non-exhaustive list of proteins associated with vertebrate junctions is shown. Proteins, which have so far been identified exclusively in vertebrate organisms, are highlighted in bold character.
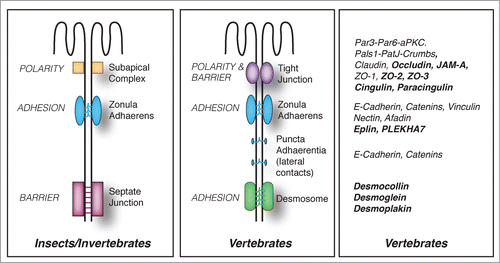
Figure 2. Proteins implicated in the organization and junctional anchoring of cytoskeletal filaments. For each type of cytoskeletal filament (actin and microtubules) the proteins shown are involved either in their polymerization, bundling, and anchoring to junctions, based on biochemical and/or cell biological evidence.
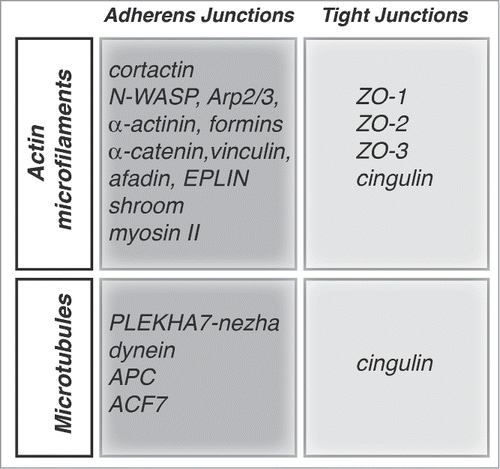
Figure 3. Crosstalk between junctions and Rho GTPases during the biogenesis of epithelial junctions. Simplified schemes showing sequential steps in the formation and maturation of the apical junctional complex (TJ and ZA) in epithelial cells, from primordial contact (top) to mature junction (bottom), and the proteins involved. Legends for graphical objects are shown in box (top left). Green and red arrows/lines indicate activation and inhibition, respectively. The main effects of Rho GTPase regulation on cytoskeletal organization and function are summarized on the sides of each scheme. Proteins and protein interactions depicted here are derived from studies on different model systems, so they do not necessarily occur together, but are grouped in one scheme for the sake of summarizing them. See text for additional details.
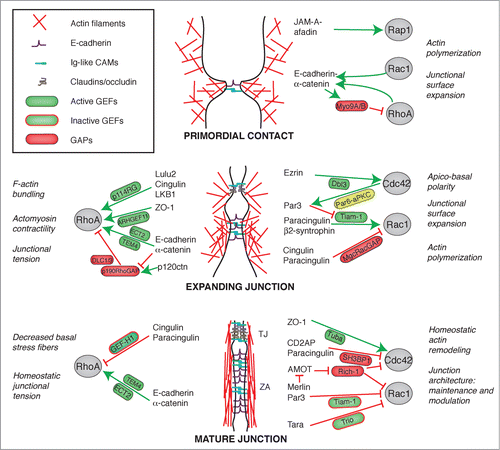
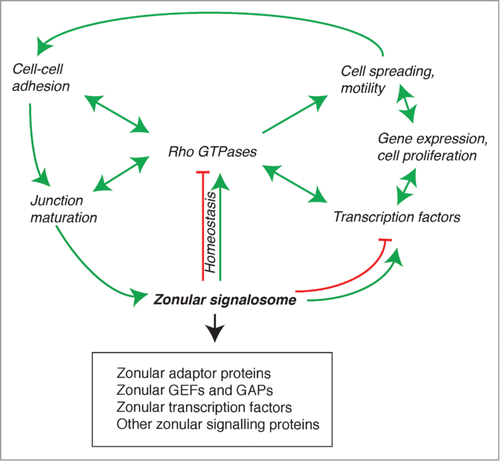
Figure 4. The zonular signalosome. The zonular signalosome is composed of zonular adaptor proteins, GEFs and GAPs, transcription factors and other signaling proteins (see text). Rho GTPases which functionally interact with the signalosome are at the center of a regulatory network that controls adhesion, junction assembly and maturation, regulation of gene expression, cell differentiation and survival, and motile behavior of cells. Transcription factors and other signaling molecules can either exist as part of the signalosome, or are cytoplasmic and regulated indirectly by the signalosome (for example, through RhoA regulation). Arrows indicate functional interactions (unidirectional or reciprocal activation, inhibition, homeostatic balance). See text for additional details.