Figures & data
Figure 1. Healthy donor CD8+ T cells stimulated in vitro with MAGE-A6172–187 and HF-2216–229 peptides cross-react to both peptides and recognize HLA-matched MAGE-A6+ tumor cells. CD8+ T cells purified from healthy donor (HD) peripheral blood underwent a single round of in vitro stimulation with the following autologous mature dendritic cell (mDC) groups: unpulsed (negative control), MAGE-A6172–187 (M6.172; M6) or HF-2216–229 (HF2.216; HF) peptide-pulsed. The relative frequencies of peptide- and tumor-specific CD8+ T-cell responders primed with the indicated peptide were measured in interferon γ (IFNγ) and granzyme B ELISPOT assays. (A) CD8+ T cells were screened for their ability to produce IFNγ in response to autologous antigen presenting monocytes pulsed with the indicated peptides, and (B) for their ability to produce IFNγ and granzyme B in response to HLA-A2+ MAGE-A6+ Mel526, SLM2, and WS-LCL cell lines. Background levels of IFNγ and granzyme B produced by CD8+ T cells stimulated with unpulsed mDC and induced by autologous monocytes or allogeneic tumor cell targets were subtracted from the presented peptide- and tumor-specific responses, respectively. Spot counts greater than 10 over the control were considered to be antigen-specific (dotted lines in (A) and (B) represent the cut-off) and the number of reactive spots per 105 CD8+ T cells are shown. Eight healthy HLA-A2+ donors (HD) were evaluated. Note: for HD5, insufficient numbers of cells were obtained after MAGE-A6172–187 stimulation to allow for testing of peptide cross-recognition; for HD2, insufficient cell numbers were obtained after peptide stimulations to allow for testing of WS-LCL recognition. Unmanipulated ELISPOT data presented in this figure are presented in Table SI and Figure S2.
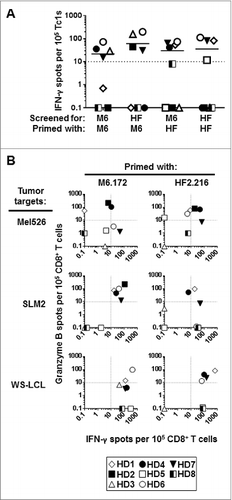
Table 1. MAGE-A6172–187 and HF-2216–229 long peptides encode for HLA-A2-restricted homologous epitopes
Figure 2. Short homologous MAGE-A6176–185 and HF-2220–229 epitopes are recognized by CD8+ T cells stimulated with the corresponding long peptide variants. Homologous MHC Class I-binding epitopes within these peptides (MAGE-A6176–185 and HF-2220–229; ) were identified using the SYFPEITHI epitope prediction algorithm. The peptides were chemically synthesized and tested for HLA binding and immunogenicity. (A) MAGE-A6176–185 (M6.176) and HF-2220–229 (HF2.220) binding affinities for HLA-A2 were determined using the T2-based HLA-A2-binding assay. T2 cells were loaded with titrated amounts of peptide (≤50 μg /mL, as indicated). Cells were stained with anti-HLA-A2 antibody and fluorophore-conjugated secondary. Mean fluorescence index (MFI) was determined by fluorescence cytometry. (B) CD8+ T cells purified from healthy donor (HD) peripheral blood primed with long peptides and evaluated in were screened for their ability to recognize autologous monocytes ± short M6.176 (sM6) and HF2.220 (sHF) using the interferon γ (IFNγ) ELISPOT assay. Data presented were adjusted for non-specific IFNγ production by CD8+ T cells, and spot counts greater than 10 over control were considered to be antigen-specific (dotted line represents the cut-off). (C-D) To further confirm the immunogenicity of the MAGE-A6176–185 and HF-2220–229 peptides, the peptides were compared to their long counterparts for their ability to sensitize CD8+ T cells against naturally-processed MAGE-A6. (C) MAGE-A6 protein expression of immature dendritic cells (iDCs) transduced with AdV.LacZ (DC.LacZ) or AdV.MAGE-6 (DC.MAGE-A6; MOI 250) shown by Western blot. Recombinant MAGE-A6 (rMAGE) was used as the positive control. Results are reflective of data obtained using 2 healthy donors. (D) CD8+ T-cells, which underwent a single round of in vitro stimulation with peptide-pulsed mature dendritic cells (mDC), were tested against autologous DC.LacZ or DC.MAGE-A6 using the IFNγ ELISPOT assay. Data are representative of 3 independent experiments. Mean ± SEM values are presented; statistical analyses were performed by single-tailed Student's T test; p < 0.05.
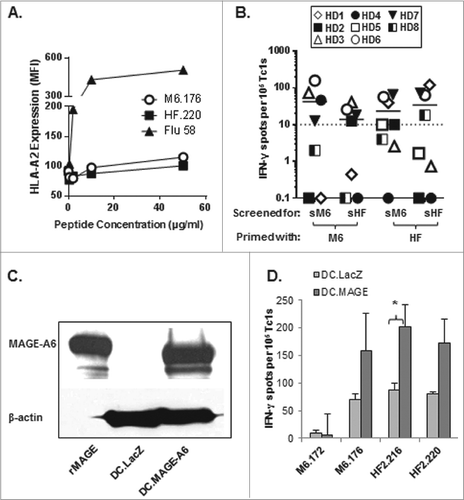
Figure 3. CD8+ T cell memory responses to MAGE-A6172–187 and HF-2216–229 long peptides. (A) Healthy donor (HD) or melanoma (Mel) patient-derived CD8+ T-cell memory responses were evaluated against MAGE-A6172–187 (M6.172) or HF-2216–229 (HF2.216) peptides. CD8+ T cells purified from fresh healthy donor or melanoma patient PBMCs were co-cultured with autologous monocytes and 10μg/mL peptides for 1.5 h, followed by 10ug/mL brefeldin A for an additional 3.5 h. CD8+CD3+cytokine+ populations were evaluated by immunofluorescence staining and flow cytometry. Non-specific cytokine responses were subtracted from the presented peptide-specific responder percentages. Median percentages of peptide-specific cytokine responders are reported in interferon γ (IFNγ) vs. IL-10 dot-plots. Note: for HD12, insufficient numbers of cells were obtained to allow for testing of HF-2216–229-specific memory responses. ((B) and C) Overall CD8+ T-cell cytokine responses against MAGE-A6172–187 or HF-2216–229 peptide, and CD45RO staining on cytokine-positive cells from healthy donors were also measured. CD8+ T cells isolated from 7 healthy donor PBMCs were tested for production of multiple cytokines by intracellular staining and CD45RO expression levels via fluorescence cytometry. (B) Median percentages of peptide-specific cytokine-positive responders are shown for TNF, IFN-γ, IL-10, and IL-2. (C) Percentage of CD45RO+ memory cells among IFNγ+ or IL-10+ CD8+ T-cell responders. Median percentage values are shown;statistical analyses were performed by single-tailed Student's T test; *p < 0.05.
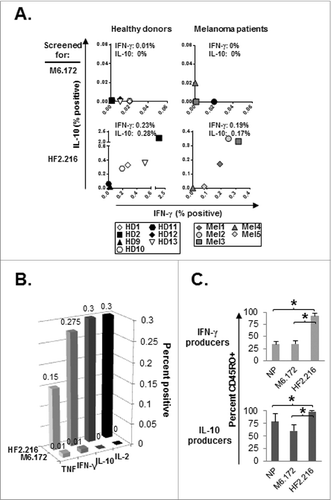
Table 2. HLA-A2+ healthy donor and melanoma patient demographic and clinical characteristics.
Figure 4. Functionality of MAGE-A6172–187 or HF-2216–229-stimulated CD8+ T-cell responders from advanced-stage melanoma patients. MAGE-A6172–187 or HF-2216–229 peptide-stimulated CD8+ T cells from melanoma patients were examined for their ability to recognize HLA-A2+, MAGE-A6+ tumor cell lines in vitro as described in . Spot counts greater than 10 over control were considered to be antigen-specific (dotted lines represent the cut-off).
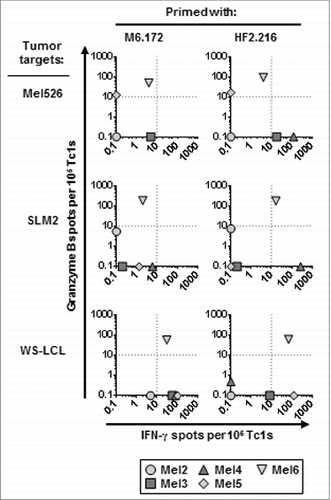
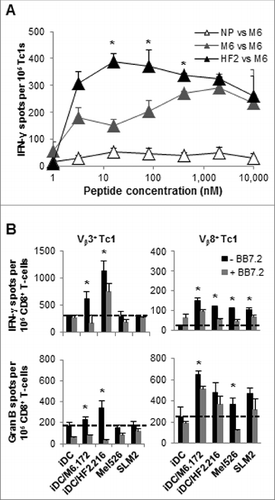
Figure 5. HF-2216–229 peptide stimulation induces high avidity, polyclonal CD8+ T cells capable of recognizing naturally-processed MAGE-A6 in an HLA-A2–restricted manner. In (A), to test the functional avidity of MAGE-A6172–187 (M6) and HF-2216–229 (HF2) peptide-primed CTLs, lymphocytes were evaluated for their ability to recognize titrated doses of the MAGE-A6172–187 peptide pulsed autologous HLA-A2+ iDC via interferon γ (IFNγ) ELISPOT assay. MAGE-A6172–187, MW = 1,772 g/mol (e.g.,, 10 μmol/L = 17.8 μg/mL); HF-2216–229, MW = 2,115 g/mol (e.g., 10 μmol/L = 21.2 μg/mL). Data from HD3 are shown, and are reflective of 2 independent experiments. NP (not pulsed with peptide) mature dendritic cells (mDC) were used for control in vitro stimulation. (B) MAGE-A6172–187–specific Vβ3+ and HF-2216–229-specific Vβ8+ T-cell clones from donors HD3 and HD12, respectively, were evaluated for their ability to recognize unpulsed (iDC), MAGE-A6172–187 (iDC/M6.172)– or HF-2216–229 (iDC/HF2.216)–pulsed autologous iDC, as well as the Mel526 and SLM2 melanoma cell lines in IFNγ and granzyme B ELISPOT assays. Analysis of 104 T effector cells and 104 control or peptide-pulsed antigen presenting cells (APCs) co-incubated in each assay well. The data presented are adjusted to represent the frequency of responders per 105 CD8+ T cells. BB7.2 anti-HLA-A2 antibody (10μg/mL) was added to replicate wells to confirm the HLA-A2-restriction of T-cell responses. Mean ± SEM values are presented. Statistical analyses were performed relative to BB7.2 blocking control by single-tailed Student's T test; *p < 0.05.