Figures & data
Table 1. Patient characteristics in HMGB1 baseline groups
Figure 1. HMGB1 protein levels in serum correlate with survival and associate with changes in tumor effusion HMGB1. (A) Serum HMGB1 concentration at baseline was measured by ELISA and plotted against overall survival (total N = 202). Patients with lower HMGB1 serum levels at baseline tended to show prolonged survival. The dotted line indicates the median HMGB1 baseline level, which was used as a cutoff for study grouping. Data are shown with offset axes to clarify survival of patients with undetectable serum levels of HMGB1. (B) Patients with lower than median HMGB1 baseline level showed significantly prolonged survival as compared to high HMGB1-baseline patients (p = 0.008, n = 101 per group, Log-Rank test). While all patients had serum available, eight patients also had tumor-related ascites or pleural effusion allowing HMGB1 measurement: (C) Serum and ascites/pleural effusion samples collected on the same day before and after oncolytic adenovirus treatment were analyzed by HMGB1 ELISA. Colon (C164), pancreatic (H339), lung (K117), mesothelioma (M126 and M158), ovarian (O26), and cervical (X331) cancer patients all had corresponding changes in intracavitary fluids and serum HMGB1 levels, with only one exception (ovarian O314 patient), suggesting a close association between circulating and tumor level changes in HMGB1. Left y-axes present the serum HMGB1 levels (solid lines) while the right y-axes indicate ascites/pleural fluid concentrations (dotted lines) that were constantly higher, as expected for local HMGB1 production at the tumor.
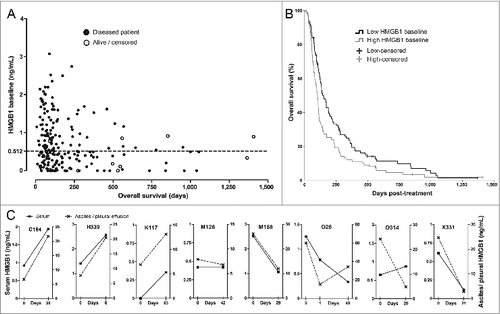
Table 2. Oncolytic adenovirus treatments and outcomes in HMGB1 baseline groups
Table 3. Multivariate analysis for disease control and overall survival
Figure 2. Antitumor T-cell activity in blood correlates with improved survival in HMGB1-low, but not in HMGB1-high patients. Antitumor T-cell activity in peripheral blood was measured by interferon-γ ELISPOT and correlated with overall survival of (A) 60 low-baseline and (B) 69 high-baseline HMGB1 patients. The longest surviving patients among (A) the low HMGB1-baseline group were those who had both induction and decrease in antitumor T-cells in their blood, which have been suggested compatible with cell amplification and trafficking to target tissues respectively (p = 0.075 as compared to “Anergy," Log-Rank test), whereas in (B) the high HMGB1-baseline group the antitumor T-cell activity did not seem to correlate with survival. (C) Patients with induction of antitumor T-cells in blood were compared based on their HMGB1 baseline status, but without significant difference (p = 0.111, Log-Rank test). When a decrease in antitumor T-cell counts in blood, a phenomenon compatible with trafficking of T-cells into tumors, was studied together with induction, (D) the low HMGB1-baseline patients had significantly improved survival as compared to high-baseline patients (p = 0.043, Log-Rank test). In panels A and B, respectively, n = 9 and 11 in “induction and decrease,” n = 19 and 25 in “induction,” n = 19 and 20 in “decrease,” and n = 13 both in “anergy;” In panel C, n = 28 in low and n = 36 in high HMGB1-baseline group; In panel D, n = 9 in low and n = 11 in high HMGB1-baseline group.
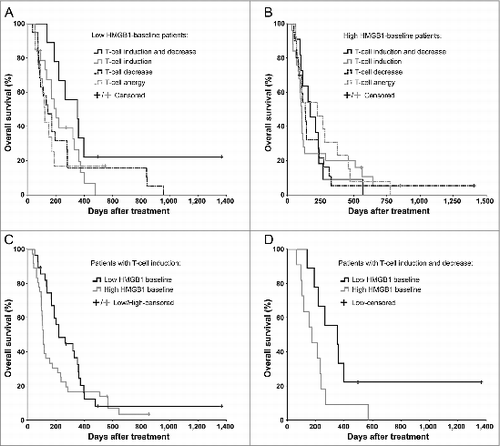
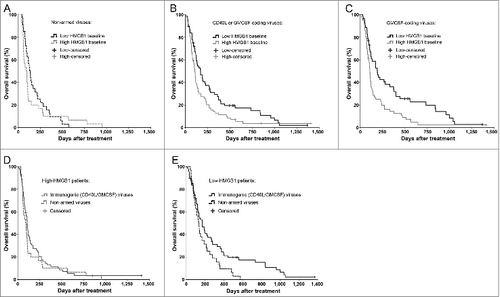
Figure 3. Immunogenic transgene coding viruses improve the survival of low-HMGB1 patients only. To test if serum HMGB1 baseline status plays even greater role in highly immunogenic treatments using oncolytic adenoviruses armed with granulocyte-macrophage colony stimulating factor (GMCSF) or CD40-ligand (CD40L), only patients with a particular first-treatment virus class were selected. Kaplan–Meier analysis based on HMGB1 baseline status revealed (A) no survival difference between the groups when treated with non-armed, less-immunogenic viruses (p = 0.315, Log-Rank test), while (B) a clear improvement in survival was observed in favor for HMGB1-low patients when treated with immunogenic transgene (CD40L or GMCSF) coding viruses (p = 0.016, Log-Rank test) and even more so with (C) GMCSF-coding viruses (p = 0.004, Log-Rank test). When comparing treatment-virus classes within the HMGB1 baseline groups, (D) high-HMGB1 patients did not show any survival differences between the virus types, while (E) low-HMGB1 patients treated with immunogenic transgene coding viruses showed a significant survival advantage (p = 0.042, Log-Rank test). Panel A, n = 33 in low, and n = 30 in high HMGB1 group; panel B, n = 68 in low and n = 71 in high HMGB1 group; panel C, n = 53 in low and n = 57 in high HMGB1 group. In panels D and E, respectively, n = 30 and n = 33 in “non-armed viruses” group, and n = 71 and n = 68 in “immunogenic (CD40L/GMCSF) viruses” group. CD40L, CD40-ligand; GMCSF, granulocyte macrophage-colony stimulating factor.