Figures & data
Figure 1. Human mammary carcinomas contain tumor stromal cells expressing metalloproteases and their inhibitors. Representative pictures of mammary cancer patient tissue array immunostaining for the different matrix metalloproteases (MMPs) and tissue inhibitors of metalloproteases (TIMPs) analyzed in breast cancer patients (200X), both at tumor center and at invasive front. (A) MMP-1, (B) MMP-2, (C) MMP-7, (D) MMP-9, (E) MMP-11, (F) MMP-13, (G) MMP-14, (H) TIMP-1, (I) TIMP-2 and (J) TIMP-3.
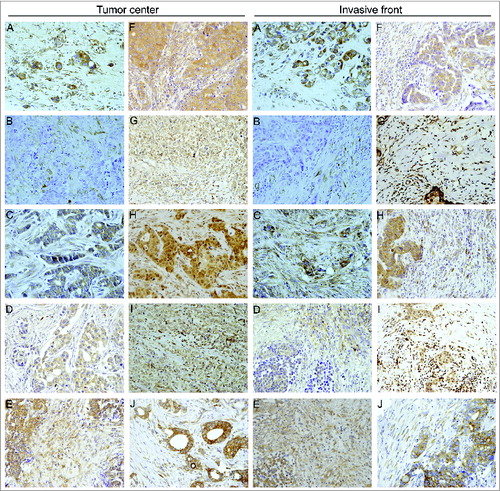
Figure 2. Human mammary carcinomas contain CAFs and MICs expressing metalloproteases and their inhibitors. Representative pictures of double-immunostaining of mammary cancer patient tissue arrays to confirm expression of matrix metalloproteases (MMPs) and tissue inhibitors of metalloproteases (TIMPs) by each type of stromal cell. (A) Positive MMP-11 staining (brown) in mononuclear inflammatory cells (MICs) identified by the CD45 marker (pink) (200X) and (B) Positive TIMP-2 staining (brown) in cancer-associated fibroblasts (CAFs) identified by the α-SMA marker (pink) (200X).
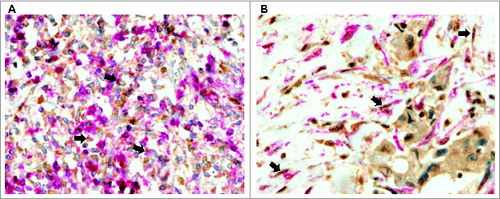
Figure 3. Prognostic significance of stromal expression of metalloproteases and their inhibitors in mammary carcinoma patients. Kaplan–Meier survival curves for relapse-free survival as a function of matrix metalloprotease 11 (MMP-11) expression by mononuclear inflammatory cells (MICs) at the tumor center (A) or at the invasive front (B), or as a function of tissue inhibitors of metalloprotease 2 (TIMP-2) expression by cancer-associated fibroblasts (CAFs) located at the tumor center (C) or at the invasive front (D).
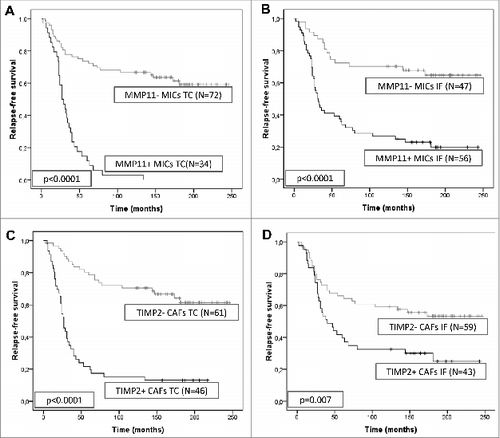
Table 1. Basal demographics and clinical characteristics of 107 patients with invasive ductal carcinoma of the breast included in our study.
Figure 4. Clustering analysis of human mammary carcinoma patient expression of metalloproteases and their inhibitors. Hierarchical clustering analysis of global expression patterns of matrix metalloproteases (MMPs) and tissue inhibitors of metalloproteases (TIMPs) among different cells types present in breast tumor specimens (n=107) as measured by immunohistochemistry on tissue microarrays from the tumor center and the invasive front. Graphical representation of hierarchical clustering results in cancer-associated fibroblasts (A, B) and mononuclear inflammatory cells (C, D). Rows: tumor samples; columns: MMPs/TIMPs. Protein expressions are depicted according to a color scale: red, positive staining; green, negative staining; gray, missing data. Two major clusters of tumors (F1A and F1B) were shown in fibroblast-like cells both in the tumor center (A), and (F2A and F2B) in the invasive front (B). For mononuclear inflammatory cells (MICs), we found 2 major cluster of tumors (M1A and M1B) in the tumor center (C), whereas 3 distinct clusters of tumors (M2A, M2B and M2C) were found at the invasive front (D).
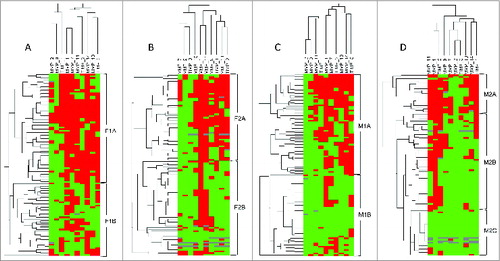
Table 2. Expression of MMPs and TIMPs in 3 different cluster groups resulting from the combination of stromal cells in the tumor center and in the invasive front. Data are expressed as number of positive cases (%).
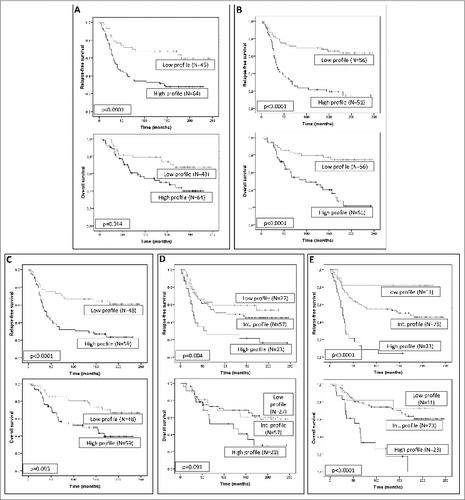
Figure 5. Survival analysis of human mammary carcinoma patients stratified according to expression of metalloproteases and their inhibitors. Kaplan–Meier survival curves (relapse-free survival and overall survival) as a function of the 2 major clusters of tumors (high and low profile of matrix metalloproteases (MMPs) and tissue inhibitors of metalloproteases (TIMPs) expression) in fibroblast-like cells at the tumor center (A) and at the invasive front (B), or in mononuclear inflammatory cells (MICs) at the tumor center (C) or at the invasive front (D). Survival curves as a function of the combination of the different cluster subgroups both in fibroblast cells and MICs, in tumor center and at the invasive front (E).