Figures & data
Table 1. Binding avidities of CrossMab for HER2-ECD
Figure 1. Characterization of Tras-Per CrossMab. (A), schematic diagram of the Fab domain exchange resulting in the generation of Tras-Per bispecific antibody when combined with the KiH technology. (B), sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS/PAGE) analysis of CD20-Flex BiFP. Samples of trastuzumab, pertuzumab, and Tras-Per CrossMab were assessed by SDS/PAGE analysis under non-reducing conditions. (C), competitive binding assay. Trastuzumab, pertuzumab, and Tras-Per CrossMab were evaluated for their ability to compete with Alexa Fluor 488-labeled trastuzumab or Alexa Fluor 488-labeled pertuzumab for binding to BT-474 cells. (D), MTS assay examining the proliferation effects of 100 nmol/L of control IgG, trastuzumab, pertuzumab, trastuzumab combined with pertuzumab, or Tras-Per CrossMab on breast cancer cell BT-474 or in the absence or presence of ErbB ligand (EGF or HRG). Results are shown as percentage of control cell proliferation. Error bars, SD. *p < 0.05. (E), in the absence of ligands, cell death induced by HER2 antibodies (10 μg/mL) were assessed by staining with SYTOX® Red and FCM. *p < 0.05. (F), in vitro ADCC analysis of trastuzumab, pertuzumab, trastuzumab in combination with pertuzumab, or Tras-Per CrossMab. PBMCs (effector cells) were added with BT-474 cells (target cells) into 96-well plates containing anti-HER2 antibodies at different concentrations. All experiments were performed in triplicate. (G), mice with BT-474 xenograft tumors were treated for the duration of the study with control IgG, pertuzumab, trastuzumab, trastuzumab + pertuzumab or Tras-Per CrossMab (2 mg/kg). Points, mean tumor volume (mm3) (n = 10 mice/group); bars, SD. *p < 0.05, Mann–Whitney test.
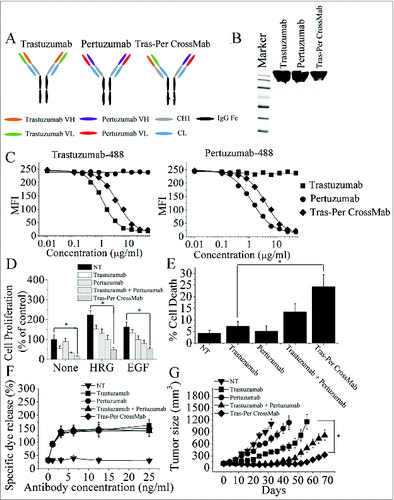
Table 2. Binding avidities of Her2 antibody variants for HER2-ECD
Figure 2. The correlation of trastuzumab and pertuzumab binding avidity-improvement and their antitumor activities. (A), antigen binding activities of trastuzumab and pertuzumab variants on SK-BR-3 cells were determined by FCM. (B), MTS assay examining the proliferation effects of 100 nmol/L of trastuzumab variants or pertuzumab variants in breast cancer cell BT-474 with the absence or presence of ErbB ligand (EGF or HRG). Results are shown as percentage of control cell proliferation. Error bars, SD. (C), in the absence of ligands, cell death induced by trastuzumab variants or pertuzumab variants were assessed by staining with SYTOX® Red and FCM. (D), In vitro ADCC analysis of trastuzumab and pertuzumab variants with binding avidity improvement. In the absence of ligands, PBMCs were added with BT-474 cells into 96-well plates containing trastuzumab or pertuzumab variants. Data are mean ± SD of at least three experiments.
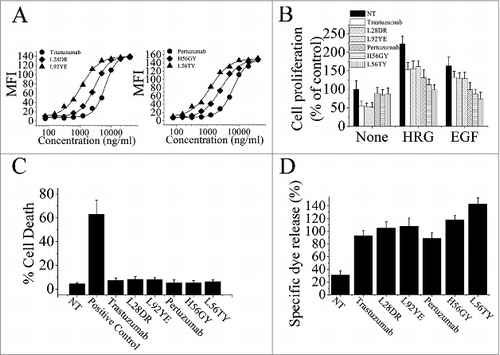
Figure 3. The in vitro antitumor effects of Tras-Permut CrossMab. (A), the anti-proliferation effect of Tras-Permut CrossMab (100 nmol/L) was evaluated by MTS assay. Mean ± SD (n = 3). *p < 0.05. (B), induction of cell death by Tras-Permut CrossMab (10 μg/mL) in the absence of ligands was assessed by staining with SYTOX® Red and FCM. The graphs are representative of at least three experiments, each of which showed similar results. *p < 0.05. (C), ADCC activity against BT-474 cells using human PBMCs as effector cells at E/T ratio of 25:1. The ADCC activity of Tras-Permut CrossMab was measured using a standard LDH assay as described in “ADCC assays.” Data are mean ± SD (n = 3). *p < 0.05.
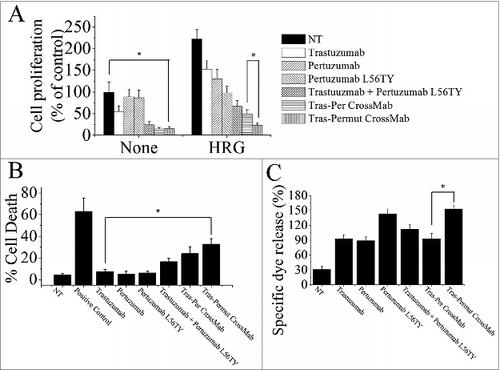
Figure 4. The therapeutic effect of of Tras-Permut CrossMab in breast cancer (BT-474) xenograft tumor models. Mice with BT-474 xenograft tumors were treated for the duration of the study with high dosage (10 mg/kg), (A) or low dosage (2 mg/kg), (B) of control IgG, trastuzumab, pertuzumab, pertuzumab L56TY, trastuzumab + pertuzumab L56TY, Tras-Per CrossMab or Tras-Permut CrossMab. Points, mean tumor volume (mm3) (n = 10 mice/group); bars, SD. *p < 0.05, Mann–Whitney test. (C), the ADCC activity of Tras-Permut CrossMab was measured using a standard LDH assay as described in “ADCC assays.” Data are mean ± SD (n = 3). *p < 0.05. (D), the role of Fc part of HER2 antibodies in their therapeutic effects. Tumor-bearing mice were treated for the duration of the study with low dosage (2 mg/kg) of trastuzumab, trastuzumab Fc mutation, pertuzumab L56TY, pertuzumab L56TY Fc mutation, Tras-Permut CrossMab and Tras-Permut CrossMab Fc mutation. Points, mean tumor volume (mm3) (n = 10 mice/group). Data are shown as means ± SEM. *p < 0.05, Mann–Whitney test.
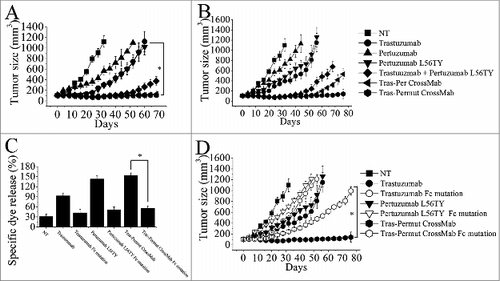
Figure 5. The calreticulin exposure induced by Tras-Permut CrossMab is essential for induction of antitumor T cell immunity against breast cancer. (A), the inhibition of cell death by caspase inhibitor Z-VAD-fmk in the range from 50 μM to 0.4 μM were evaluated after 5 d (in the absence of ligands). Mean ± SD (n = 3). *p < 0.05. (B), the surface exposure of CRT was determined by immunofluorescence cytometry 48 h after treatment with HER2 antibodies. The antibody-untreated group stained with an anti-CRT antibody was used as the negative controls. *p < 0.05. (C), in vivo antitumor protection depends on CRT. 4T1-HER2 cells were transfected with the indicated siRNAs, then treated with rCRT and/or Tras-Permut CrossMab. The antitumor response was measured by challenging BALB/c mice simultaneously with Tras-Permut CrossMab-treated tumor cells in one flank and untreated, live tumor cells in the opposite flank (n = 20). The siRNA-transfected cells were measured by immunoblotting. (D), BALB/c mice were inoculated with Matrigel-mixed 4T1-HER2 cells and then treated with Tras-Permut CrossMab or control Ig (2 mg/kg). After the first inoculation, the CrossMab-treated mice were secondarily inoculated with Matrigel-mixed 4T1-HER2 cells in the opposite flank (n = 25). Some mice were treated with anti-CD4 mAb, anti-CD8 mAb, anti-CD4 and anti-CD8 mAbs or isotype Ig. Naive mice were inoculated with Matrigel-loaded 4T1-HER2 cells as the control. *p < 0.05, Mann–Whitney test. (E), after injection of siCRT-treated 4T1-HER2 cells (containing 0.1mL Matrigel) into the inguinal mammary fat pads of female BALB/c mice, the mice were treated with Tras-Permut CrossMab (2 mg/kg). After the first challenge, the CrossMab-treated mice were subsequently inoculated with Matrigel-mixed 4T1-HER2 cells in the opposite flank (n = 25). Some mice were treated with rCRT and/or anti-CD4 and anti-CD8 mAbs (n = 25). Naive mice were inoculated with Matrigel-loaded 4T1-HER2 cells as the control. The primary tumor material is examined through measurement of tumor size. *p < 0.05, Mann–Whitney test.
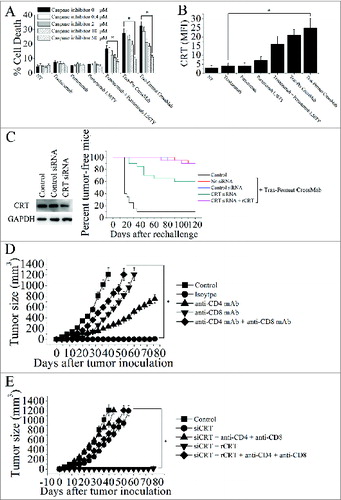
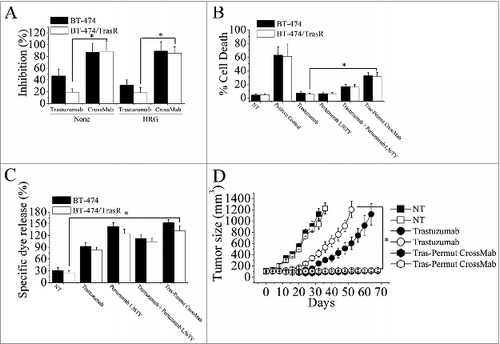
Figure 6. Combating trastuzumab-resistance by Tras-Permut CrossMab. (A), MTS assay examining the proliferation effects of Tras-Permut CrossMab (100 nmol/L) in both trastuzumab-sensitive and -resistant breast cancer cells (BT-474 and BT-474/TrasR) in the absence or presence of ErbB ligand (HRG). Results are shown as percentage of cell proliferation inhibition. Error bars, SD. Tras-Permut CrossMab was abbreviated as CrossMab. *p < 0.05. (B), Induction of cell death against trastuzumab-resistant breast cancer was assessed by staining with SYTOX® Red and FCM (In the absence of HER2 ligands, the concentration of antibodies is 10 μg/mL). Data are mean ± SD (n = 3). *p < 0.05. (C), ADCC activities of trastuzumab, pertuzumab L56TY, trastuzumab in combination with pertuzumab L56TY, or Tras-Permut CrossMab against both trastuzumab-sensitive and -resistant breast cancer. Mean ± SD values from four separate experiments. *p < 0.05. (D), tumor volume of trastuzumab-sensitive and -resistant BT-474 breast tumor xenografts after treatment with control IgG (10 mg/kg), trastuzumab (10 mg/kg), or Tras-Per CrossMab (10 mg/kg). Data are shown as means ± SEM. *p < 0.05, Mann–Whitney test.