Figures & data
Figure 1. Partner protein interactions of eIF4G in higher eukaryotes. (A) The RNA and protein binding domains in wheat eIF4G (TaeIF4G), eIFiso4G (TaeIFiso4G), human eIF4G (HseIF4G), and yeast eIF4G (SceIF4G) are shown. Interaction domains for partner proteins are indicated by color with a key included. Domains for yeast eIF4G are defined as described previously.Citation37 (B) Below the N-terminal PABP binding site of wheat eIF4G is a sequence logo for the region corresponding to the PABP interaction site in human eIF4G. For this analysis, a sequence alignment from 6 monocot and 7 dicot eIF4G homologs was generated using EBI MUSCLE. The aligned sequences were submitted to WebLogo Berkeley to generate a sequence logo, in which the most conserved one, 2, or sometimes 3 residues at each position are shown as the consensus. Below the protein sequence logo are the sequences of the PABP interaction sites for human eIF4G1 and eIF4G2 as well as the corresponding sequence for wheat eIF4G. In each case, residues conserved with the consensus sequence are colored as in the sequence logo. The logo was constructed from eIF4G homologs from Triticum aestivum (wheat, JN091779), Brachypodium distachyon (Bd1g25002), Oryza sativa (Os07 g36940), Setaria italica (Si028648), Panicum virgatum (Pv00019592 and Pv00063738), Phaseolus vulgaris (010G043700), Ricinus communis (29709), Brassica rapa Chiifu-401 (Bra014505), Thellungiella halophila (Thhalv10005736), Capsella rubella (Cr10016570), Arabidopsis thaliana (AEE80028), and Arabidopsis lyrata (939058).
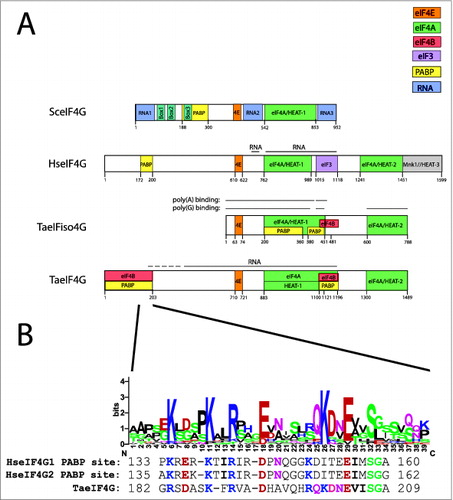
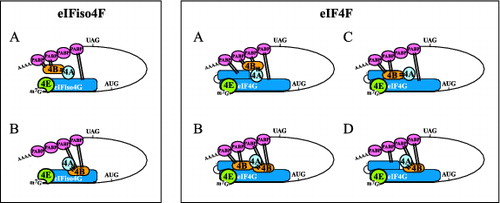
Figure 2. Mutually exclusive binding of PABP and eIF4B to eIFiso4G and eIF4G in plants. For eIFiso4F: (A) binding of PABP or (B) binding of eIF4B is depicted. For eIF4F: (A) binding of PABP to both PABP binding sites, (B) binding of eIF4B to both eIF4B binding sites, (C) binding of eIF4B and PABP to the N- and C-terminal binding sites, respectively, or (D) binding of PABP and eIF4B to the N- and C-terminal binding sites, respectively, is depicted. eIF4B is shown interacting with eIF4A (indicated with bars) but whether eIF4B can bind eIF4A when bound to eIFiso4G or eIF4G is unknown.
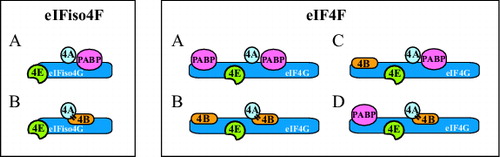
Figure 3. Partner protein interactions of eIF4B in higher eukaryotes. (A) The RNA and protein binding domains in wheat eIF4B (TaeIF4B), human eIF4B (HseIF4B), and yeast eIF4B (SceIF4B) are shown. Interaction domains for partner proteins are indicated by color with a key included. The 7 repeats in yeast eIF4B represent 26 amino acid repeats implicated in promoting translation.Citation59 (B) Below the wheat eIF4B is a sequence logo for the region encompassing the PABP and eIF4A binding site repeats and the RNA binding domain that separates the repeats. A sequence alignment from 16 monocot and 11 dicot eIF4B homologs was generated using EBI MUSCLE. The aligned sequences were submitted to WebLogo Berkeley to generate a sequence logo, in which the most conserved one, 2, or sometimes 3 residues at each position are shown as the consensus. Below the protein sequence logo is the consensus sequence for the region in which residues conserved with the sequence logo are similarly colored. The logo was constructed from eIF4B homologs from Triticum aestivum, Brachypodium distachyon (Bradi5g13500 and Bradi3g47700), Oryza sativa (Os04 g40400 and Os02 g38220), Setaria italica (Si009805 and Si016924), Sorghum bicolor (Sb06 g020170 and Sb04 g024860), Zea mays (GRMZM2G163471, GRMZM2G066815, and GRMZM2G139614), Panicum virgatum (Pavirv00022707 and Pavirv00022708, Pavirv00021798, and Pavirv00059810), Phaseolus vulgaris (l002G259400), Ricinus communis (29792), Brassica rapa Chiifu-401 (Bra026941), Thellungiella halophila (Thhalv10007279 and Thhalv10003946), Capsella rubella (Carubv10008811 and Carubv10016953), Arabidopsis thaliana (AT1G13020 and AT3G26400), and Arabidopsis lyrata (920166 and 484402).
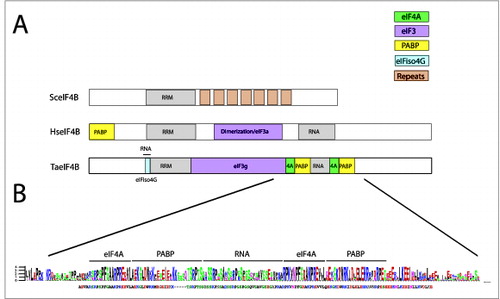
Figure 4. Partner protein interactions of PABP in higher eukaryotes. (A) The RNA and protein binding domains in wheat PABP (TaPABP), human PABP (HsPABP), and yeast PABP (ScPABP). are shown. Interaction domains for partner proteins are indicated by color with a key included. (B) Below the wheat PABP is a sequence logo for higher and lower eukaryotic PABP proteins encompassing the eIF4G, eIFiso4G, and eIF4B binding sites of the wheat PABP. For this analysis, a sequence alignment from yeast, animal, and plant PABP homologs was generated using EBI MUSCLE. A sequence logo was generated from the aligned sequences in which the most conserved one or residues at each position are shown as the consensus. The sequences part of RRM1 or RRM2 domains are indicated by the labels above the logo. The RNP2 motif of RRM2 is indicated by the asterisks. The fourth β sheet (β4) of RRM1 and the first β sheet (β1) of RRM2 are indicated by the labels below the logo. The logo was constructed from PABP homologs from Saccharomyces cerevisiae (ScPABP, NM_001179055), Schizosaccharomyces pombe (SpPABP, NM_001018809), Homo sapiens (HsPABP, NM_002568), Xenopus laevis (XlPABP, NM_001086735), Mus musculus (MmPABP, NM_008774), Physcomitrella patens (PpPABP, Pp1s257_72V6), Arabidopsis thaliana (AtPABP, At2g23350), and Triticum aestivum (TaPABP, TAU81318). Below the logo are the sequences from each species used in which invariant residues are colored as in the sequence logo.
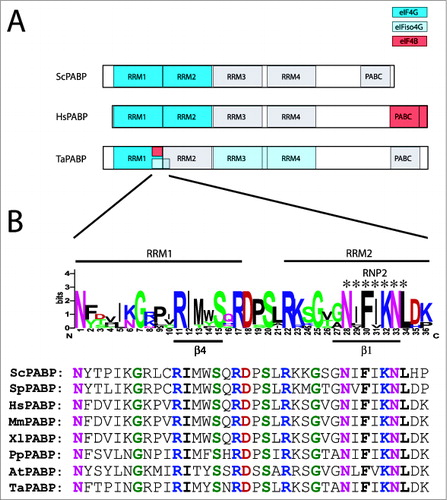
Figure 5. Models for possible interactions between PABP and eIFiso4G, eIF4G, and eIF4B during translation initiation in plants. For eIFiso4F: (A) The interaction between the termini of a translating mRNA is facilitated by interactions between PABP to eIFiso4G and eIF4B (indicated with bars). eIFiso4G contains a single binding site for PABP and this interaction is depicted. However, because eIF4A binds eIF4B (which contains 2 eIF4A binding sites), an interaction between eIF4B and 2 molecules of PABP is also depicted. Whether eIF4B can bind 2 molecules of PABP simultaneously and whether eIF4A can bind eIFiso4G and eIF4B simultaneously is unknown. (B) The interaction between the termini of a translating mRNA is facilitated solely by the binding of PABP to eIF4B which in turn is bound to eIFiso4G which contains a single eIF4B binding site. However, because eIF4B contains 2 binding sites for PABP, it is depicted as interacting with 2 molecules of PABP. For eIF4F: (A) The interaction between the termini of a translating mRNA is facilitated through the binding of PABP at the N- and C-terminal PABP binding sites in eIF4G. Because eIF4A binds eIF4B, an interaction between eIF4B and 2 molecules of PABP is also depicted. (B) The interaction between the termini of a translating mRNA is facilitated solely through the binding of PABP to eIF4B which is bound at the N- and C-terminal eIF4B binding sites in eIF4G. Because eIF4B can dimerize, the 2 molecules of eIF4B are shown as a dimer but whether dimerization occurs during binding to eIF4G is unknown. (C) The interaction between the termini of a translating mRNA is facilitated through the binding of 2 molecules of PABP to eIF4B bound at the N-terminal eIF4B binding site in eIF4G and the binding of PABP to the C-terminal PABP binding site in eIF4G. (D) The interaction between the termini of a translating mRNA is facilitated through the binding of PABP to the N-terminal PABP binding site in eIF4G and the binding of 2 molecules of PABP to eIF4B bound at the C-terminal eIF4B binding site in eIF4G.