Figures & data
Figure 1. LC3 and EDEM1 colocalize with EAV-induced dsRNA foci. (A) Vero E6 cells were infected with EAV at a multiplicity of infection of 1 TCID50/cell and fixed at 16 h p.i before being processed for immunofluorescence analysis using antibodies against dsRNA and either PDI, LAMP1, LC3 or EDEM1. Insets show enlargements of the boxed areas. (B) Summary statistics of the samples shown in (A) expressed as the percentage of dsRNA puncta colocalizing with the indicated marker protein signals. Error bars represent the standard deviations from counting 50 cells in 3 independent experiments.
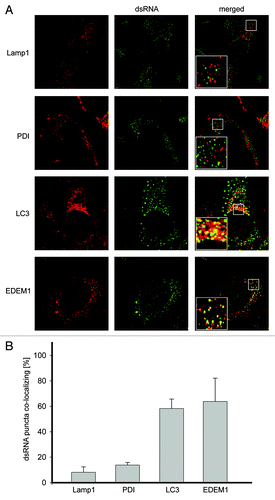
Figure 2. EAV infection induces the formation of dsRNA- and LC3-positive DMVs. (A) BHK-21 cells infected with EAV or mock treated were chemically fixed at 16 h p.i. and embedded with Epon resin. Numerous DMVs associated with ribosomes were found mostly in the perinuclear region of the EAV-infected cells (left panel). Asterisks indicate some of the DMVs. (B) BHK-21 cells (mock)-infected with EAV and fixed at 16 h p.i. were processed for IEM. Cryo-sections were labeled with antibodies against dsRNA (top right panel), LC3 (top or bottom left panel) or both (bottom right panel). Arrowheads indicate some of the DMVs. Insets show examples of labeled vesicles at high magnification. The size of the gold particles is indicated in the image double labeled sample. ER, endoplasmic reticulum; M, mitochondria; N, nucleus. Scale bars: 200 nm.
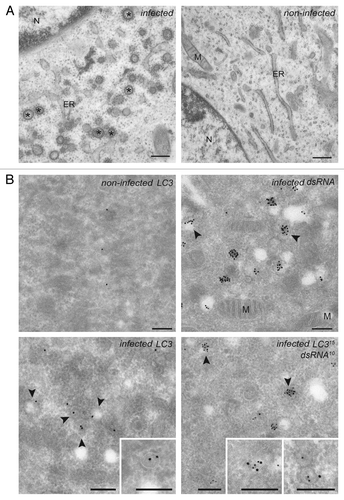
Figure 3. EAV replication and association of LC3 with the DMVs does not depend on an intact autophagy machinery. (A) End point 10-fold dilutions of an EAV stock were titrated on Atg7+/+ and atg7−/− MEFs. Cytopathic effects were scored at 4 d p.i. and TCID50 units per ml were calculated. Similar titers were observed, indicating that Atg7 deletion does not affect EAV entry, replication or egress. Values presented in the graph are calculated and expressed as the log10 of TCID50 units per ml of supernatant, and the plotted values represent the average of 3 experiments. Standard deviations are indicated. (B) The Atg7+/+ and atg7−/− MEF cells were infected with EAV and fixed and processed for immunofluorescence analysis as described in Materials and Methods using antibodies against LC3 and dsRNA.
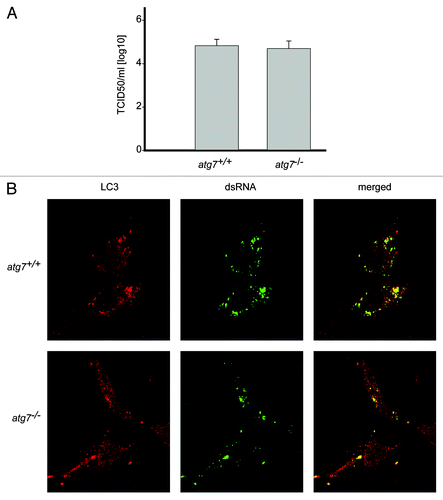
Figure 4. EAV-induced DMVs are associated with LC3-I but not with N-terminally GFP-tagged LC3. Vero E6 cells were transfected with plasmids expressing either C-terminally HA-tagged nonlipidated LC3-I (LC3-HA) (A) or N-terminally GFP-tagged LC3 (GFP-LC3) (B), and subsequently infected with EAV for 16 h before being processed for immunofluorescence analysis.
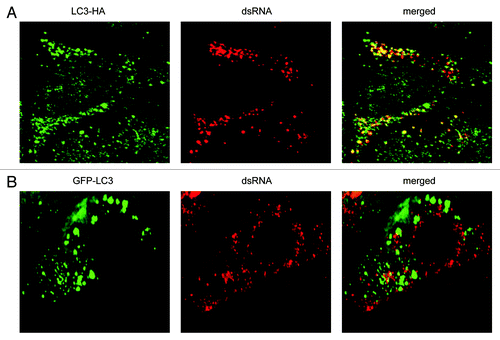
Figure 5. LC3 is required for EAV life cycle. Vero E6 cells were transfected with siRNAs directed against LC3A and LC3B (siRNALC3) or with control, scrambled siRNAs (siRNAscr). At 48 h post transfection, cells were inoculated with EAV. (A) Cells (mock-) infected with EAV for 16 h were lysed and protein extracts were processed for western blot analysis using antibodies recognizing LC3, N protein or tubulin. Tubulin was used as a loading control. (B) End point 10-fold dilutions of an EAV stock were titrated on the indicated transfected cells. Cytopathic effects were scored at 3 d p.i. and TCID50 units per ml were calculated. (C) Immunofluorescence examination using antibodies against the indicated proteins. Dotted lines highlight the cell contours.
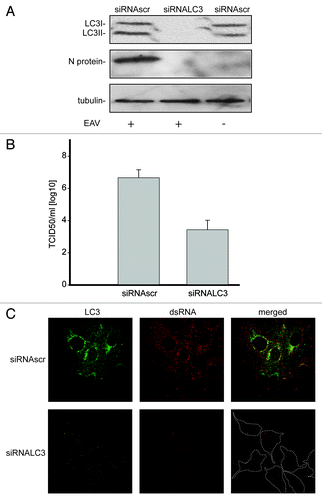
Figure 6. Nonlipidated LC3 restores EAV replication in LC3-depleted cells. (A) Vero E6 cells were cotransfected with siRNA directed against LC3A and LC3B (siRNALC3) and the plasmid expressing nonlipidated LC3-HA, which is not targeted by the siRNA probes. Cells were then infected with EAV at 48 h post transfection and fixed at 16 h p.i. before being processed for immunofluorescence analysis using antibodies against the indicated proteins. (B) Statistical analysis of the experiment shown in (A). The graph illustrates the percentage of cells, positive or negative for LC3-HA that contained dsRNA puncta, i.e., productively infected by EAV.
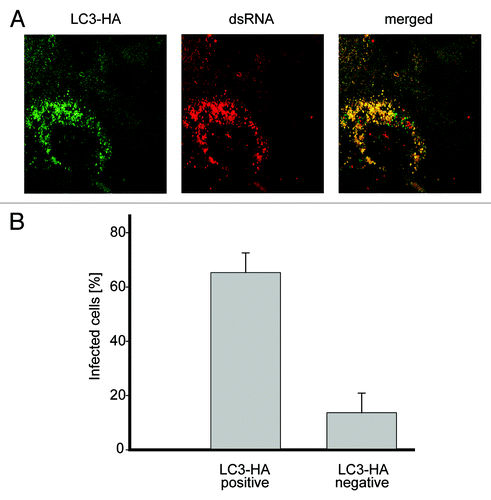
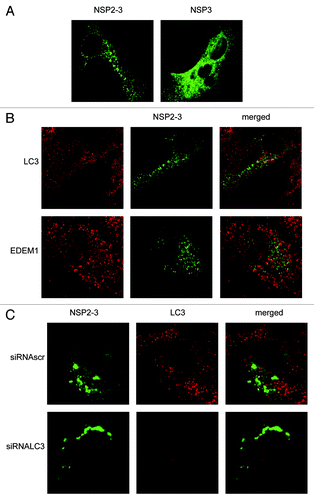
Figure 7. EAV NSP2-3–induced membrane rearrangements do not colocalize with LC3 and are formed independently of LC3. Vero E6 cells were infected with vTF7-3 and subsequently transfected with plasmids expressing GFP fusion proteins with either EAV NSP2-3 or NSP3. Cells were fixed at 5 h p.i. and processed for immunofluorescence analysis. (A) Comparison of the subcellular distribution of NSP2-3 and NSP3. (B) Cells expressing NSP2-3 were immuno-stained using antibodies against LC3 or EDEM1. (C) Cells were transfected with siRNAs directed against LC3A and LC3B (siRNALC3) or control siRNAs (siRNAscr) 48 h prior to infection with vTF7-3. Cells were processed for immunofluorescence analysis using the LC3-specific antibodies.