Figures & data
Figure 1 Interactions of UP9C and LSU proteins. (A) The pJK1 and pJK2 plasmids contain UP9C in the BD- and AD-vectors, respectively. The yeast strains co-transformed with indicated BD- and AD-plasmids were grown on selective media without leucine and tryptophan (SD-LT), leucine, tryptophan and adenine (SD-LTA), leucine, tryptophane and histidine (SD-LTH) or were screened for β-galactosidase expression (+X-gal). (B) The protein gel blot shown on the left-hand side verifies expression of the GST-fusion proteins in the extracts from bacteria producing GST-ZZ (2b), GsT-Joka8 (8) and GST-Joka20 (20); the GST-PB1 protein was not detected. The protein gel blot shown on the righthand side shows results of the “pull-down” assay performed as described in Materials and Methods. The results confirm interaction of UP9c with Joka8, Joka20 and the ZZ domain of Joka2; the extract containing His-tagged UP9C protein (C+) was loaded as a positive control and the arrows indicate the positions of the proteins corresponding to the expected sizes of the recombinant proteins: GST-ZZ (54.5 kDa), GST-Joka8 (66.5 kDa), GST-Joka20 (42 kDa) and His-UP9C (17.2 kDa). (C) The scheme of the protein (truncated NpJoka2) encoded by the insert present in pJoka2. The position of EcoRI used for the subcloning is shown and the domains PB1 and ZZ are indicated. (D) The pJK11 plasmid contains the insert as pJoka2 but in pGAD424. Plasmids pLUs1–4 contain the corresponding orfs (LSU1–4 from Arabidopsis) cloned into pGBT9. The remaining explanations as in (A).
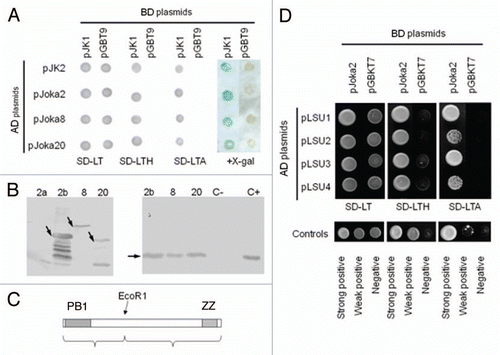
Figure 2 The family of p62/NBR1/Joka2 proteins. (A) The phylogenic tree was constructed using full-length protein sequences by the parsimony methods and 100 bootstrap replicates using SEQBOOT, PROTPARS and CONSENS of the Phylip v.3.69 program package. The bootstrap values are given at the respective branches. The three subfamilies are marked and identified by the name of a typical member (p62, NBR1 or Joka2). The accession numbers of the proteins used in the analysis are shown in Table S1. (B) characteristic domains present in the p62, NBR1 and Joka2 proteins from Nicotiana tabacum, Arabidopsis thaliana and Homo sapiens. Proteins and domains are drawn to scale. See text for details and domains description.
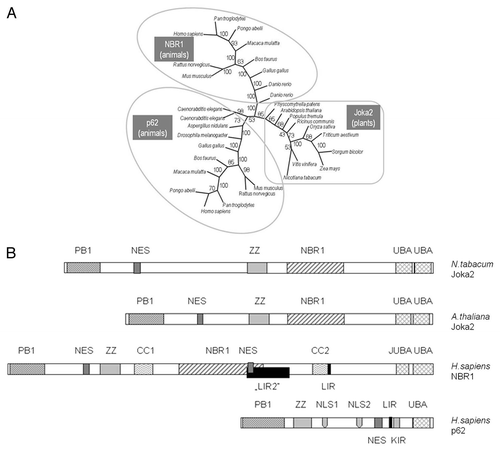
Figure 3 Interactions of Joka2 and ATG8f. (A) The protein gel blot with anti-GST (α-GST) and anti-His (α-HIS) antibodies showing the results of the “pull-down” experiment using the mixture of crude exctracts prepared from the bacteria producing His-ATG8f and GST-Joka2. After affinity purification through the Ni column, both proteins are detected (lane 3) due to ATG8f-Joka2 interaction. The controls, without GST-Joka2 and without His-ATG8f, are shown in lanes 1 and 2, respectively. The negative controls with the GST-tag (produced from the empty vector pGEX4T-1) and his-ATG8f protein and with the GST-Joka2 protein and the His-tag (produced from the empty vector pET28a) are shown in lanes 1 and 2, respectively. For α-GST the high and the low range proteins are shown in the upper part and the middle part, respectively. The molecular marker (M) with the size (kDa) of the proteins is indicated. (B) The results of Y2H experiment and (C) the scheme of the Joka2 fragments present in the used plasmids. AD means fusions of the activating domain of GAL4 with the indicated protein or domain. BD means fusions of the DNA binding domain of GAL4 with the indicated protein or domain. Pluses and minuses indicate the growth and lack of growth, respectively, on the plate shown on the lefthand side. The growth is an indicator of protein-protein interaction. Proteins and domains shown in (B and C) are drawn to scale.
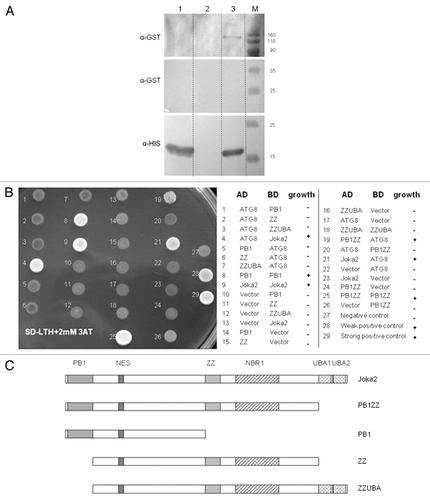
Figure 4 Co-localization of Joka2-YFP and CFP-NtATG8f in Nicotiana benthamiana grown in sulfur-deficient medium for 2 d before agroinfiltration. The parts show accumulation of CFP-NtATG8f fluorescence signal (CFP-NtATG8f), accumulation of NtJoka2-YFP fluorescence signal (Joka2-YFP), overlay of both (Merge) and the corresponding tissues under bright field (Light). The white arrow indicates the site of signals co-localization. Scale bars (10 µm) are shown.

Figure 5 Localization of Joka2-YFP in tobacco seedlings. (A) The overlays of the signal for Joka2-YFP, DNA staining with DAPI, chlorophyll signal (observed as a red fluorescence of the chloroplasts; seen only in green parts) and brightfield image are shown for the indicated numbers of days post sawing (dps) in the indicated parts of the J4-1 seedlings grown in the indicated conditions (-S, sulfur starvation; -N, nitrogen starvation; nS, nutrient sufficient medium and water). Images 1–4 are magnifications of the indicated parts. Notice, a difference in the number of green spots between the roots (Joka2-YFP abundant) and the shoots (Joka2-YFP hardly detected), which is clearly observed in the shoot-root transition zone (magnified picture #3). Arrows point the cytosolic spots, while arrowheads the nuclei. (B) The signals of DAPI-stained nuclei, fluorescent protein (Joka2YFP for J4-1 or eGFP for AB5), red signal after acridine orange staining (AO) and the overlay of all is shown for the indicated plants: LA Burley 21 (LAB21) and transgenic AB5 (producing eGFP) used as a control and J4-1 grown in water for 14 d. extranuclear Joka2-YFP speckles and red spots stained by AO (if present) are shown by arrows. Arrowheads point the nuclei. (C) The treatment with protease inhibitor E64d increases the number of spots including Joka2-YFP. Both upper parts show the Joka2-YFP signal, while both lower parts show the corresponding tissues in the transparent view (Light). The scale bars are indicated. The separate signals used for the overlay are shown in Figures S2–5.
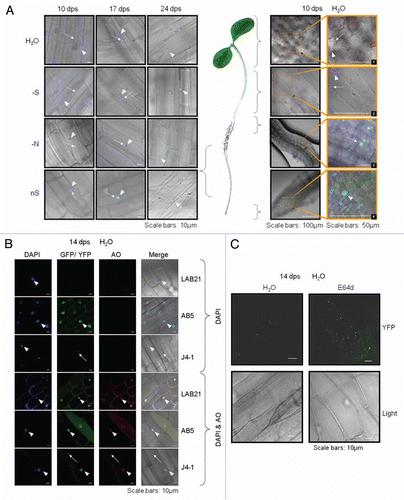
Figure 6 Expression of Joka2, ATG8f and UP9 in various parts of LA Burley 21 plants. The 8-week-old plants grown in nutrient-sufficient (nS) medium were transferred for 2 d into S- or N-deficient medium and, as a control, to nS again. Gene expression was monitored by sqRT-PCR. expression of actin (Tac9) served as a control. The UP9 contains the mixture of UP9C and UP9A (both induced by sulfur starvation) because the used primers amplify both products.
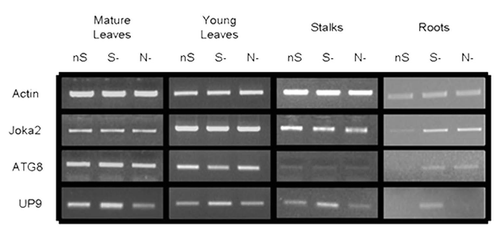
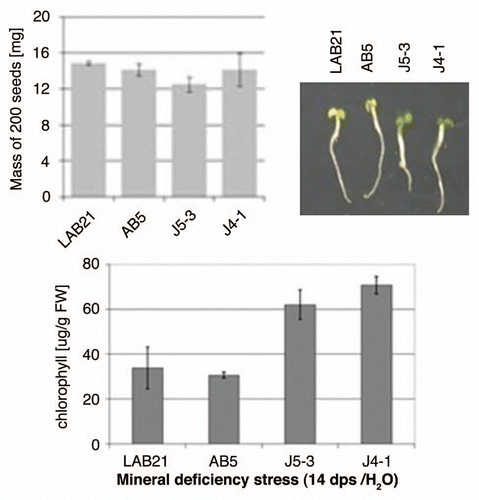
Figure 7 Effect of mineral deficiency stress on plants overproducing Joka2 fusion proteins. The mass of 200 seeds determined before sowing as well as the phenotype and chlorophyll content of tobacco seedlings germinated and maintained for 14 d in water are shown. LAB21, LA Burley 21; AB5, the line producing EGFP; J4-1, the line producing Joka2-YFP, J5-3, the line producing Joka2-CFP.