Figures & data
Figure 1 The effect of the ectopic expression of PML-RARα on autophagic activity in leukemic and nonleukemic cells. (A and B) U937/PR9 (A) and indicated cells (B) were respectively treated with or without 100 µM ZnsO4, or with EBSS for the indicated hours. Cell lysates were harvested for immunoblotting proteins as indicated. LC3-II protein expression was quantified according to the densitometric value and the relative folds against untreated cells were shown as means ± SD from three independent tests. (C) TEM micrographs of the indicated treatments in U937/PR9 cells. AV-like structures were indicated by arrowheads. “Nuc” stands for nucleus. The scale bars on control, EBSS- and Zn2+-treated cells were 1,000 nm. The high-magnification picture (right part of bottom row) was from the framed area in the Zn2+-treated cell (scale bar = 500 nm). (D) Quantification of data in part (C). The percentage of AV+ cells was shown in the left part and among these AV+ cells, the percentage of cells with indicated AV numbers per AV+ cell were indicated in the right part. The symbol *indicates a p value of less than 0.05 against U937/PR9 control cells. (E) U2Os cells were transfected with GFP-LC3 alone (top row) or co-transfected with GFP-LC3 and DsRed-PML-RARα, DsRed-PML or DsRed vector (bottom row). After 24 h, cells were examined by confocal microscopy. The representative images of the indicated transfected cells with the corresponding treatments were shown. The values (x ± SD) represent the percentage of GFP-LC3 puncta-positive cells from three independent experiments. The symbols * and # respectively indicated p values of less than 0.001 and 0.01 compared with the untreated cells with GFP-LC3 transfection alone. The symbol & indicated a p value of less than 0.05 compared with the cells co-transfected with DsRed and GFP-LC3 plasmids. All experiments were repeated at least three times and similar results were obtained.
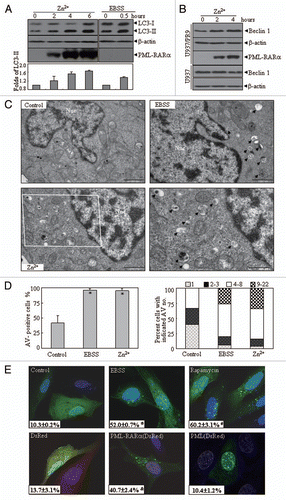
Figure 2 In vivo effect of PML-RARα expression on autophagy in leukemic mice. Leukemic cells (3 × 105) from BM and spleen of APL transgenic mice were injected into FVB/N mice via the tail vein. Animals were sacrificed about 29 d after leukemic cell transplantation. (A) Cytologic analysis by Wright's Giemsa staining of peripheral blood (PB) and BM. Histopathological sections of spleen from the indicated mice were stained with H&E. Images were observed by microscopy with a Nikon digital camera. (B) The proteins from BM cells and spleens of the normal and diseased mice were extracted and the indicated proteins were detected by protein gel blot. (C) Representative electron micrographs of myelocytes and promyelocytes in BM from normal and diseased mice were observed by TEM. The scale bars were 2,000 nm on the top images and the high-magnification picture (bottom part) was from the framed area of the middle part (scale bar = 1,000 nm). (D) The percentage of cells with the indicated AV numbers per immature granulocyte in BM from the indicated mice were calculated and summarized. All experiments were repeated three times and similar results were obtained.
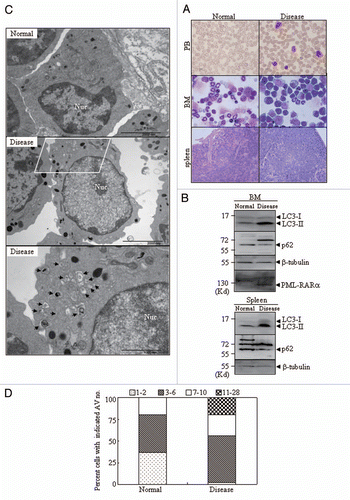
Figure 3 The effects of PLZF-RARα and NPM-RARα fusion proteins on autophagy. U2OS cells were co-transfected with GFP-LC3 and hcRed-PLZF-RARα, DsRed-PML-RARα, or the corresponding empty vectors (HcRed or DsRed), or with Myc-LC3 and CFP-NPM-RARα or the CFP vector. After transfection for 24 h, the cells were stained with anti-Myc antibody or directly analyzed by confocal microscopy. The Myc-LC3 signal was imaged on the red fluorescent protein (RFP) channel, and the CFP signal was obtained on the CFP channel. (A) Representative images of the cells transfected with the indicated constructs were shown. Arrowheads indicate cells with the expression of proteins as labeled. (B) The percentage of GFP-LC3 puncta-positive cells (left part) and the total number of GFP-LC3 dots per cell (right part) were calculated. The symbol *indicates a p value of less than 0.001 compared with the cells co-transfected with DsRed and GFP-LC3 plasmids. (C) After a transient transfection with the indicated plasmids, U2OS cells were extracted and detected by protein gel blot. The transfected expressions of APL-related fusion proteins were confirmed by a RARα antibody. (D) U937/PLZF-RARα cells were treated with 100 µM ZnSO4 for the indicated hours and the cell lysates were harvested for immunoblotting. Relative LC3-II in (C and D) was determined by the ratio of densitometric value of LC3-II relative to the corresponding empty-transfected or the untreated controls. All experiments were repeated three times with similar results, and all values were shown as means with bar as SD of three independent experiments.
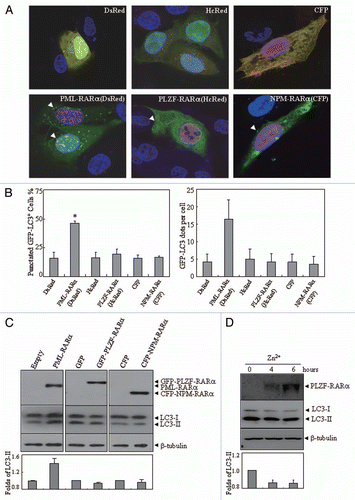
Figure 4 The effects of lysosomal enzyme inhibitors and 3-MA on altered localization and expression pattern of LC3 protein induced by PML-RARα. (A and B) U2OS cells were transiently co-transfected with GFP-LC3 and DsRed-PML-RARα (bottom parts) or DsRed (upper parts) for 24 h, followed by treatment with or without 3-MA (10 mM) and pepstatin A (10 µg/ml) plus E64d (10 µg/ml) for an additional 4 h. Then the cells were observed by confocal microscopy. The representative images for each treatment are shown (A). Quantification data of the percentage of GFP-LC3 puncta-positive cells and GFP-LC3 dots per cell are shown in the left and right parts, respectively (B). Symbol * stands for p < 0.05. (C and D) U2OS cells were transiently transfected with the indicated concentrations of Flag-PML-RARα (Flag-P-R) expression vector or the empty Flag vector (1.0 µg of each plasmid was transfected in C) for 24 h and then treated with or without pepstatin A (10 µg/ml) and E64d (10 µg/ml) (C), 3-MA (10 mM) (D) or an equal volume of the vehicle for an additional 4 h. Cell lysates were harvested and analyzed by protein gel blot with specific antibodies. Relative LC3-II expression in (C and D) was determined by the ratio of the densitometric value of LC3-II relative to the empty-transfected controls with vehicle treatment. All experiments were repeated three times with similar results, and values are shown as means with bar as SD of three independent experiments.
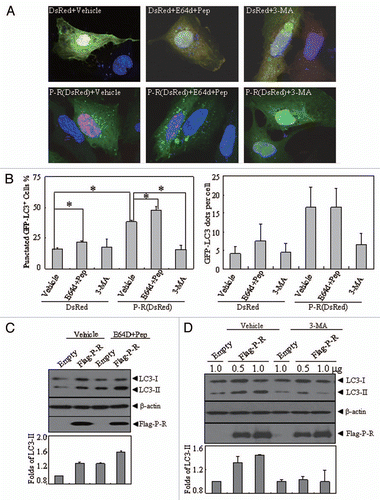
Figure 5 The effects of PML-RARα and PLZF-RARα overexpression on the Akt-mTOR pathway in leukemic cells. U937/PR9 and U937 cells (A) as well as U937/PLZF-RARα cells (B) were treated with or without 100 µM ZnSO4 for the indicated hours. Total proteins were harvested and the indicated proteins were analyzed by protein gel blot. The induced expression of PML-RARα and PLZF-RARα proteins in the respective cells were confirmed by the RARα antibody. All experiments were repeated at least three times and similar results were obtained.
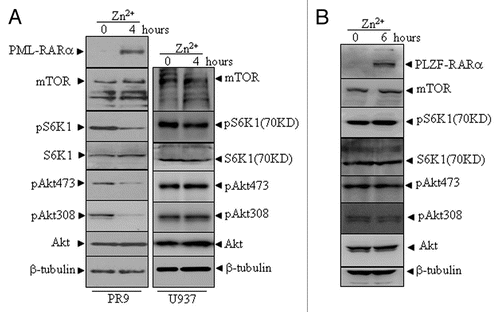
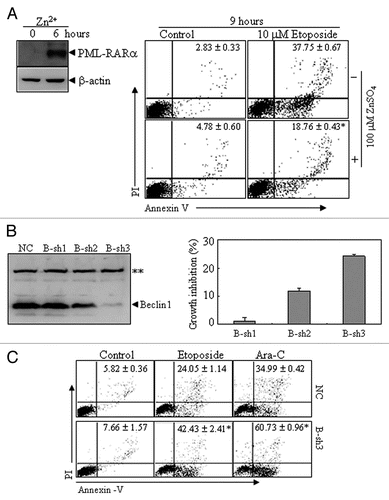
Figure 6 The effects of PML-RARα-induced autophagic activity on apoptosis in leukemic cells. (A) U937/PR9 cells were pre-treated with or without 100 µM ZnSO4 for 6 h, followed by treatment with or without etoposide. Then annexin V+ cells with or without PI staining were detected by flow cytometry. PML-RARα protein induction was also shown. (B) NB4 cells were stably transfected with B-sh1, B-sh2, B-sh3 or NC. The effect of Beclin 1 suppression by shRNAs was analyzed by protein gel blot (left part) and the percentage of growth inhibition against NC cells are shown as means ± SD from three independent experiments (right part). (C) NC- and B-sh3-expressing cells were respectively treated with or without etoposide (1.5 µM), or with Ara-C (2 µM) for 24 h. Then annexin V+ cells with or without PI staining were detected by flow cytometry. In (A and C), the values represent the percentages of the corresponding cells in means ± SD of triplicates from an independent experiment. The symbol * indicates p values of less than 0.01 compared with etoposide-treated U937/PR9 cells without Zn2+ induction (A) or NC cells with corresponding treatments (C). The symbol **points to a nonspecific band. All the experiments were repeated three times and similar results were obtained.