Figures & data
Figure 1. Multi-step phosphorylation-mediated regulation of Vav. (A) Schematic representation of the domain structures of Vav. (B) Vav activation is achieved via initial phosphorylation of Tyr-142 and Tyr-160 in the Ac region, resulting in disruption of the CH domain interaction with the PH domain. It is hypothesized that this allows for exposure of Tyr-174. Subsequent phosphorylation of this residue results in conformation change that allows RhoGTPase to access the DH domain.
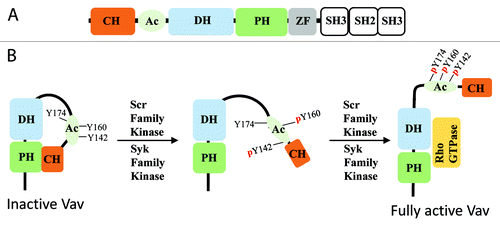
Figure 2. Domain structures and phosphorylation sites of RhoA-specific GEFs. Schematic diagram of the domain structure of RhoGEFs, and depiction of phosphorylation sites and kinases that mediate activation (green) and inhibition (red) of RhoGEFs. Kinases that are known to phosphorylate and regulate RhoGEF function where phosphorylation sites have not been identified are listed on the right (see text for detail). CC, coiled-coil; DH, Dbl homology; LR, leucine rich; NLS, nuclear localization signal; PDZ, post-synaptic density 95; disk large, zona occludens-1; PBM, PDZ binding motif; PH, Pleckstrin homology; RH, RGS homology; and ZF, Zinc finger-like binding domain.
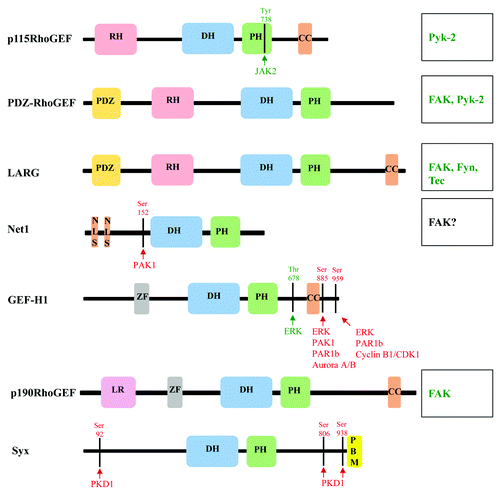
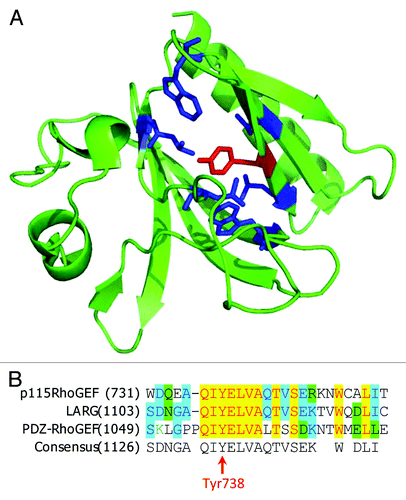
Figure 3. Phosphorylation of PH domain of p115RhoGEF by JAK2. (A) Structure of p115RhoGEF’s PH domain (PDB identifier: 3ODO). TheTyr738 residue is depicted in red and the surrounding hydrophobic residues are depicted in blue. (B) Alignment of amino acid sequences for p115RhoGEF, LARG, and PDZ-RhoGEF.