Figures & data
Figure 1 Cyclin D1 binds to PACSIN 2 through its C-terminal E-rich motif. (A) Immunoprecipitation (IP)-western blot (WB) was performed to determine the binding of endogenous cyclin D1 and Pacsin 2. Protein lysates were prepared from either NIH 3T3 cells or murine liver tissue. IP was conducted with a cyclin D1 antibody pre-conjugated to agarose. The immunoprecipitates was subjected to WB for detection of Pacsin 2. (B) IP-WB was conducted of cells transfected with either an expression vector for Myc epitope-tagged PACSIN 2 and FLAG-tagged cyclin D1 wild type or mutant expression vectors. The schematic representation shows the acidic-rich region. The C-terminal E-rich motif is required for PACSIN 2 binding. (C) Amino acid sequence of the carboxyl terminal region of cyclin D1. IP-WB of PACSIN 2 association with D-type cyclins, wild type or mutant is as shown. IP was conducted with the FLAG antibody directed to the N-terminus of cyclin Ds with western blot to detect the Myc epitope of PACSIN 2. The amino acid residues required for the binding were mapped to E278 and E280 for cyclin D1 and E272 and E274 for cyclin D2, respectively. (D) Schematic representation of constructs of D-type cyclins that were used for mapping the domain requirement for PACSIN 2 binding.
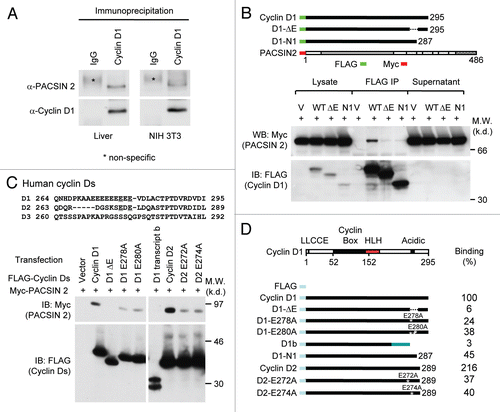
Figure 2 The NPF motifs of PACSIN 2 are required for cyclin D1 binding. (A) Schematic representation of PACSIN 1, 2 and 3. (B) The mammalian expression vectors encoding PACSIN 1, 2 or 3 were co-expressed with FLAG-tagged cyclin D1 and subsequent IP-WB was conducted. The Myc epitope of PACSINs are shown by western blot of cellular lysates. Co-precipitation of cyclin D1 with PACSIN 2 is shown by immunoprecipitation of cyclin D1 with FLAG antibody. (C and D) HEK293T cells were co-transfected with mutant constructs of PACSIN 2 (as indicated) together with vector encoding FLAG-cyclin D1. Immunoprecipitation was performed with an anti-Myc antibody following by western blot using FLAG antibody to detect cyclin D1.
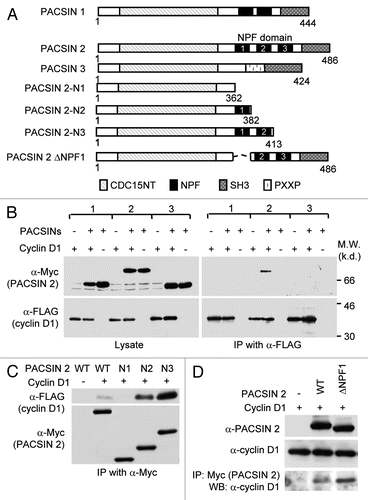
Figure 3 Knockdown of PACSIN 2 reduced cell migration. (A) Prostate cancer cells, LNCaP, were transiently transfected with PACSIN 2 siRNA (siPACSIN 2). 72 hr after transfection, western blot was conducted for PACSIN 2 protein. Actin was included as protein loading control. (B) Transwell migration assays were performed with cells transfected with either control siRNA (Ctrl. siRNA) or siPACSIN 2. (C) Quantitative analysis of trans-migrated cells. (D) 3T3 cells were assessed for cell spreading. Cells transfected with siPACSIN 2 have reduced cell spreading phenotype. (E) Quantitative analysis of cell spreading (arbitrary unit). (G and H) NIH 3T3 and LNCaP cells were transiently transfected with siPACSIN 2 or control. Western blot was performed to determine the expression of cyclin D1 and PACSIN 2. (I) MCF-7 cells were transfected vector encoding PACSIN 2. GDI and β-actin served as protein loading control.
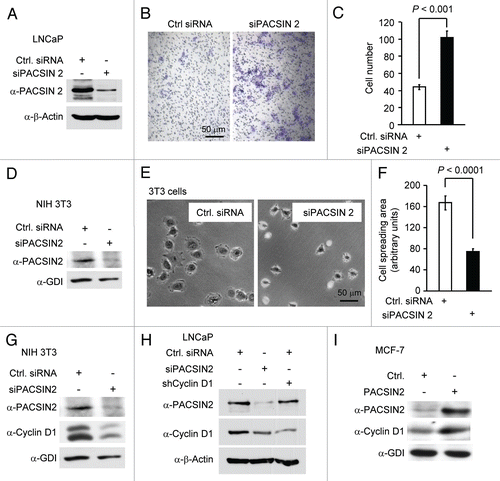
Figure 4 PACSIN 2 repression of cellular migration is dependent on cyclin D1a. (A) Cyclin D1−/− MEFs were transduced with viral expression vector (MSCV-IRES-GFP) encoding cyclin D1a, cyclin D1b or GFP control. (B) Cells were transiently transfected with siPACSIN 2. Western blot was performed with an anti-PACSIN 2 antibody to show a reduction of endogenous PACSIN 2. (C) Transwell migration assays were conducted in triplicate. Cells that migrated were stained and counted. Five fields in each of triplicate wells were randomly recorded. (D and E) The time-lapse video-microscopy was conducted in cyclin D1+/+ and cyclin D1−/− MEFs transfected with a PACSIN 2 expression or control vector. Vector expressing GFP was co-transfected. The GFP-positive cells were further analyzed for cell migration velocity. Student t-test was used for quantitative analysis of cell migration.
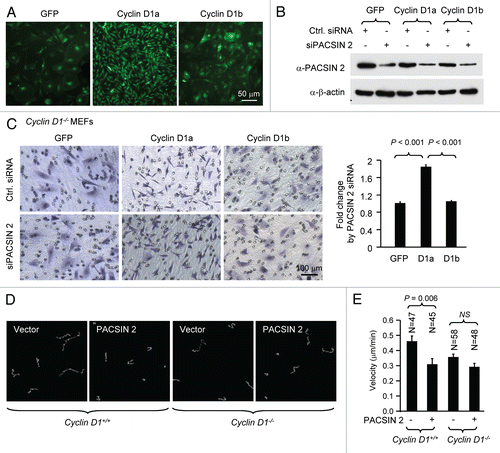
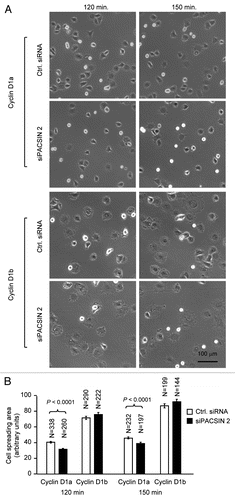
Figure 5 PACSIN 2 regulation of cell spreading depends on cyclin D1 expression. (A) Cyclin D1−/− cells transduced with either cyclin D1a or cyclin D1b retrovirus and transfected with either control or siPACSIN 2 were assessed for cell spreading at different time points after seeding. (B) Quantitative analysis of cell spreading (arbitrary unit). Student t-test was used for quantitative analysis of cell spreading. p-value and number of cells analyzed were indicated in the figure. N is for the number of cells counted. Data are representative of three separate experiments.