Figures & data
Figure 1 Temporal correspondence between DNA replication and E2F. (A) (Top) Overview of successive temporal events in DNA replication. Gene products regulated by E2Fs are shown in blue. The licensed pre-replication complex (pre-RC) contains CYCLIN E, ORC, CDC6, CDT1 and MCM situated at origins of replication (ORI). Initiation involves MYC and CYCLIN A:CDK-mediated activation of pre-RC helicase activity. Delicensing occurs through inhibition of CDT1 by protein sequestration by GEMININ along with SKP2 and PCNA-mediated ubiquitination. A temporal delay between CYCLIN E- and CYCLIN A-associated CDK activity is mediated by APC/CCDH1. APC/C, anaphase-promoting complex/cyclosome with CDH1; GEM, GEMININ; ORC, origin recognition complex; CYC, CYCLIN complexed with cyclin-dependent kinase (CDK); MCM, minichromsome maintenance proteins 2–7; Pol, DNA polymerase. (Bottom) Typical temporal pattern for E2F activators (E2F1–3) as cells re-enter the cell cycle from quiescence (G0) following growth factor stimulation. The temporal dynamics summarized in this review are indicated: (1) Delayed E2F increase relative to immediate early genes; (2) switching OFF to ON; (3) amplitude modulation and (4) switching ON to OFF. (B) (Left) Genes induced by E2F1–3 and their associated network modules. (Right) Overview of network logic involving modules of the RB-E2F network. IN, upstream signals originating from growth factor signaling and MYC.
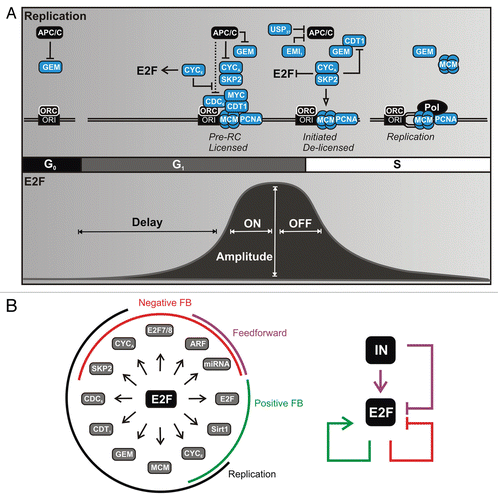
Figure 2 Switching OFF to ON: Derepression and positive feedback on E2F. (A) Events following growth factor receptor engagement that increases E2F expression. In the exit from quiescence, a repressive p130:E2F4/5 complex (designated by RBp:E2FRNuc) situated on E2F target genes is removed via phosphorylation and degradation of p130; E2F4/5 is exported from the nucleus (Nuc) to the cytoplasm (Cyt) in the absence of p130. RB, on the other hand, cycles through states of hypo- and hyperphosphorylation during the cell cycle. Repression of p53 is achieved through E2FA-mediated upregulation of the SIRT1 deacetylase. Green arrows indicate positive feedback. GF, growth factors; E2FA, activators E2F1–3; E2FR, repressors E2F4–5; RBp, hypophosphorylated pocket proteins; RBpp, hyper-phosphorylated pocket proteins; p21WAF1, cyclin-dependent kinase inhibitor; (B) Effect of positive feedback on gene expression. Cell-cell variability in isogenic cells (noise) can manifest in differences in expression dynamics. Depicted are time courses from distinct cells with a gene subject to weak (gray) or strong (black) positive feedback. Positive feedback can decrease the time needed to exceed basal expression (time to cross horizontal dotted line; τDelay) and the coherence across a population (difference in τDelay between cells).
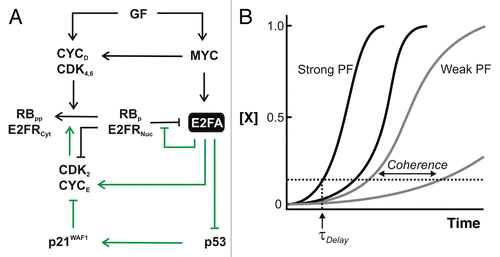
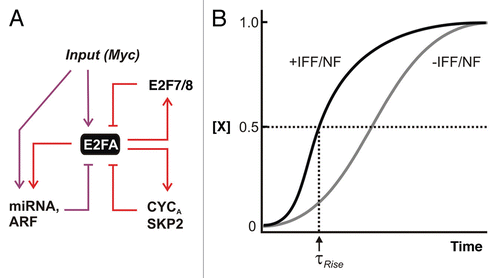
Figure 3 Switching ON to OFF—negative feedback and feedforward. (A) Negative feedback and incoherent feedforward (I1-FFL) modules. Direct regulation of miR-17–92 cluster of miRNAs along with the tumor suppressor Arf by both MYC and E2F represent incoherent feedforward/negative feedback loops. CYCLIN A and SKP2 are involved in suppression of DNA binding and stability of E2F1–3, respectively. E2f7/8 are transcriptional targets of E2F that bind to and suppress E2f1–3 transcription in the late phases of the cell cycle. (B) Effect of rapid incoherent feedforward (I1-FFL) and negative feedback (NF). Repression provided by I1-FFL/NF can reduce steady state but when offset by increased production rate, it can allow a shorter rise time (τrise) compared with a circuit without IFF/NF.