Figures & data
Figure 1 ESAP1 directly binds to EWS-FLI1. (A) ELISA type assay using bacterial expressed and secreted alkaline phosphatase conjugated peptides with direct binding to full length recombinant EWS-FLI1. Vector only expressed and secreted alkaline phophatase conjugated peptide (■ V-AP) and ESAP1 encoded and secreted alkaline phosphatase conjugated peptide (▲ ESAP1-AP) were placed recombinant EWS-FLI1 coated wells, respectively. (● V-AP and □ ESAP1-AP peptides were placed on BSA coated wells). (B) Dose-dependent competition of synthetic ESAP1 against EWS-FLI1 and ESAP1-AP complexes measured using ELISA type assay with recombinant EWS-FLI1. (■ and ▲ represent EWS-FLI1 coated wells that were treated with ESAP1-AP and 1:10 diluted ESAP1-AP peptides, respectively. ▼ represents EWS-FLI1 coated wells that were treated with vector only-AP peptide. ◆ represents BSA coated wells that were treated with ESAP1-AP peptide). (C) Binding kinetics and the affinities of Antennapedia-fused synthetic ESAP1 is examined using SPR in Biacore T100. For this representative experiment, Ka = 1.436 × 107, Kd = 2.849 and KD = 1.984 × 10−7 M were calculated. X-axis is time (s), y-axis is arbitrary real-time binding unit (response unit-RU). Chi square was 152 and U was 4 for this specific example.
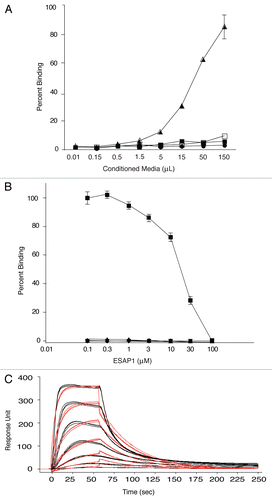
Figure 2 Lysine-rich core of ESAP1 required for cytotoxicity. (A) A dose-response experiment of FITC-tagged antennapedia fused ESAP1 (△), Antennapedia fused ESAP1 without FITC (■) and fragments (□-FITC-Antp-ESAP1+K, ○-FITC-Antp-ESAP1−K) were exposed to the TC32 cell line in monolayer growth. (B) FITC-tagged antenniapedia fused ESAP1, ESAP1+K and ESAP1−KI peptides entered TC71 cells in 20 min. Hoeschst 3342 is counter-stain for nucleus in the left column. Cellular peptide uptake is shown in middle column. Right column presents merged image of nuclear staining and FITC peptides.
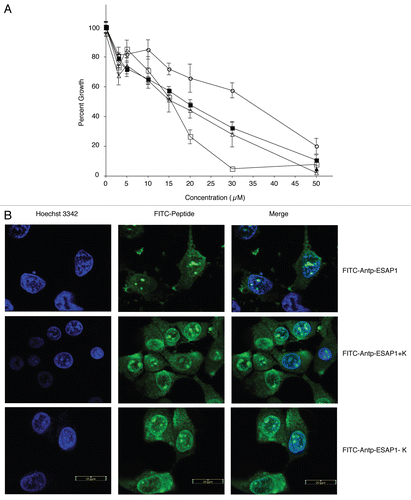
Figure 3 ESAP1 inhibits ET cell lines growth. (A) ET (▲, TC71 and ■, TC32) Type I fusion protein bearing ET cell lines, (▼, A4573) Type III fusion protein bearing ET cell line and cell lines without EWS-FLI1 (○, SKNAS-neuroblastoma and □, PANC1-Pancreas) were treated with ESAP1. Cells were incubated for 4 d followed by formazan dye-reduction assay to determine the number of viable cells. The assay was repeated between 2–4 times depending on the cell line. (B) An EGFP-ESAP1 fusion protein was expressed in ET (TC71) cells and impaired the anchorage independent growth by 4.45-fold. Fluorescent colonies were counted under an epifluorescence microscope. (C) Soft agar colony numbers significantly reduced in EGFP-ESAP1 expressing cell line (p = 0.0158).
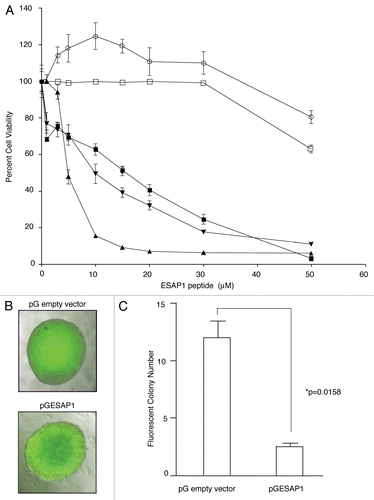
Figure 4 ESAP1 reduces EWS-FLI1 regulated transcript activation. (A) The NR0B1 promoter luciferase activity was evaluated for EWS-FLI1-dependent transcriptional activity in Cos-7 cells. (B) Exogenously expressed EWS-FLI1 levels were confirmed by immunoblotting. (C and D) GLI1 and cyclin E transcript levels were measured with qRT-PCR. mRNAs of synchronized cells (open bars) and the 8 h ESAP1-treated cells (gray bars) were isolated and subjected to the qRT-PCR with respective primers. GAPDH was used as a normalizer for fold difference calculation by 2−ΔΔCt method. Student's t-test was used to evaluate statistical significance of fold difference of the ESAP1 treatment. p values were given in the graph.
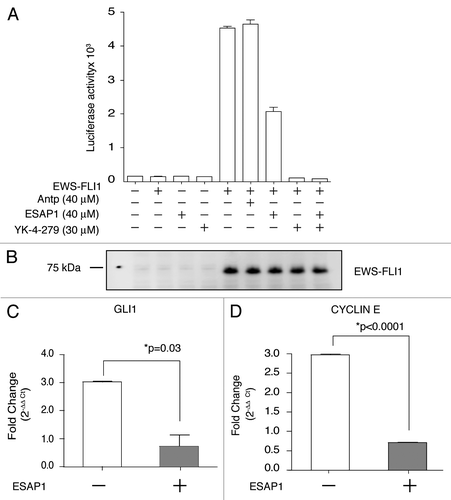
Figure 5 ESAP1 blocks cell cycle progression in a Ewing tumor cell line. Unsynchronized (A and B) or nocodazole synchronized (C and D) TC71 cells were treated with ESAP1 (B and D) or untreated in order to evaluate cell cycle kinetics. Cells were harvested at times indicated and treated with propidium iodide followed by flow-cytometric analysis. Cell phase percentages calculated in ModFit software. This experiment was representative of four repetitions. (E–H) The light microscope with 20x final magnification captured the representative morphologic appearance of the cells at the 24 h of the treatment respective to cell cycle evaluation. Sizing bar is equal to 100 µm. (I–L) Cell cycle histograms showed the examples of the DNA content aberrations. (M) Cell cycle real time PCR array results. Cellular mRNA was analyzed from the 8-h time point of ESAP1 treatment. The fold differences of mRNA expression were calculated with the five different housekeeping gene's averages for each gene using a 2−ΔΔCt method.
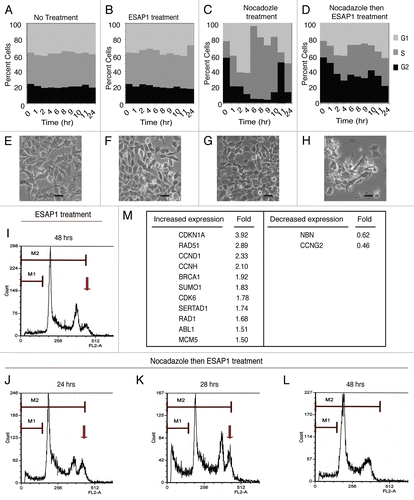
Table 1 Ewing sarcoma antagonist peptide sequences revealed in phage display
Table 2 KD values of peptides measured in SPR (µM)