Figures & data
Figure 1 Depletion of PA28γ enhances cellular sensitivity to radiomimetic treatment. Left part: clonogenic survival curves of CAL51 cells transfected with siRNA against PA28γ for 96 h and subsequently treated with various concentrations of the radiomimetic drug NCS. Cells transfected with siRNAs against ATM or GFP served as controls. The experiment was performed in triplicates. Right part: protein gel blotting analysis showing the extent of protein knockdown. Total cellular extracts of CAL51 cells transfected with the various siRNAs for 96 h were blotted with the indicated antibodies.
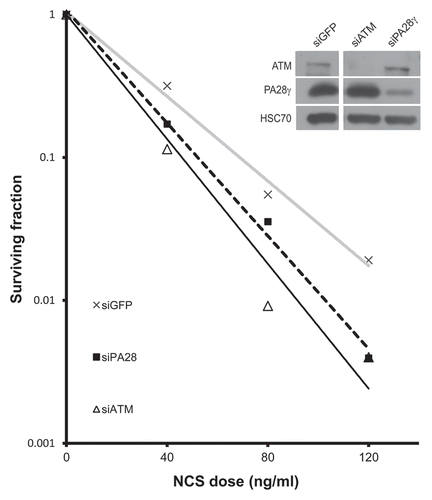
Figure 2 Depletion of PA28γ affects the disappearance of DNA damage-induced nuclear foci of γH2AX, 53BP1, MDC1 and BRCA1. CAL51 cells were treated with 10 ng/ml NCS 72 h after transfection with siRNAs against PA28γ or GFP, fixed and stained with antibodies against γH2AX (A), 53BP1 (B), MDC1 (C) and BRCA1 (D). (A, C and D) The number of cells in which more than 10 foci were counted. Mean of three independent experiments is presented, and error bars represent SD (*p-value < 0.05; **p-value < 0.01; ***p-value <0.0005, by χ2 analysis). (B) The mean of 53BP1 foci per cell is presented, and error bars represent standard error (*p-value < 0.05; ***p-value < 0.0005; by student's t-test). Experiments were performed in triplicates. (E) Protein gel blotting analysis of total cellular extract of CAL51 cells 72 h after siRNA transfection, showing the extents of protein knockdown in this experiment.
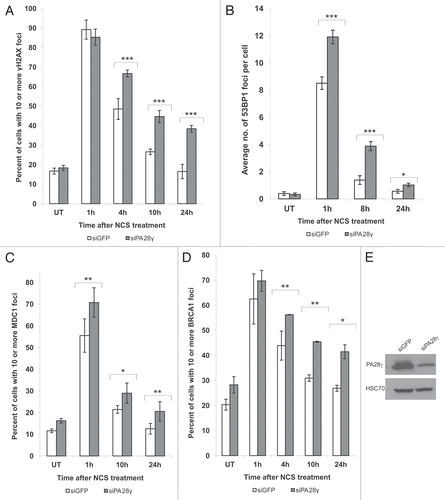
Figure 3 Deficiency of DSB repair in PA28γ-depeleted cells. (A) Direct observation of DNA damage 2 h after treatment of U2OS cells with 200 ng/ml of NCS using a neutral comet assay. (B) Quantitation of the comet data. The length and intensity of SYBR green-stained DNA tails relative to heads is shown as the relative comet tail moment (n = 100). p-values indicate the statistical significance of the difference between samples (student's t-test). Bars represent standard error of the mean based on two independent experiments. (C) Protein gel blotting analysis showing the extent of PA28γ knockdown.
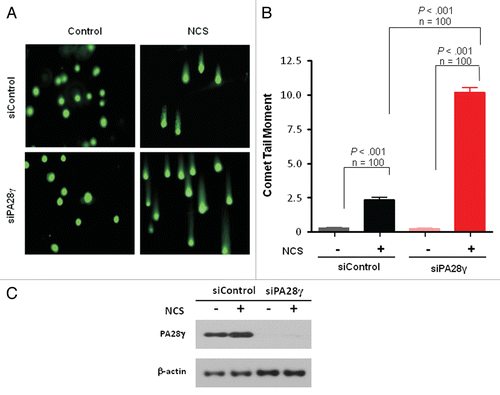
Figure 4 Effect of PA28γ depletion on the NHEJ and HRR pathways of DSB repair. The experimental systems are based on cells in which interrupted GFP-encoding sequences containing recognition sites of the rare cutter restriction endonuclease I-SceI were incorporated into the cellular genome. In one system, the repair of I-SceI-induced DSB via HRR regenerates an active GFP-encoding sequence,Citation69,Citation71 and in the other, this effect is obtained following NHEJ-mediated repair of the break.Citation70 In both cases, the GFP signal is monitored using FACS. (A) Effect of PA28γ depletion on NHEJ. HeLa cells containing the reporter sequences for NHEJCitation39,Citation70 were transfected with an I-SceI-encoding plasmid along with the indicated siRNA oligonucleotides and analyzed 72 h later by flow cytometry. GFP-positive cells are gated, and the percentage of GFP-positive cells in PA28γ-depleted cells is normalized against that of cells transfected with irrelevant siRNA (siLuciferase). Cells depleted of the KU70 protein, a major NHEJ player, served as a positive control. Shown is the mean of the NHEJ ratio (average of triplicates). Error bars represent standard error. Results of one of three independent experiments are shown. (B) Similar analysis in U2OS cells containing the HRR reporter.Citation69 Cells depleted of the RAD51 protein, a major HRR player, served as positive control. Results of one of four independent experiments are shown. Error bars represent standard deviation. (C) Quantification of the accumulation of RAD51 at DSBs in the presence or absence of PA28γ. U2OS cells were transfected with siRNAs against luciferase (siControl) or PA28γ, cultured for 48 h and irradiated with α-particles as described in references Citation40 and Citation68 to produce linear tracks of DSBs. The effectiveness of the downregulation was analyzed by immunoblotting, as displayed on the right. At the indicated time points after irradiation, the cells were stained for DNA (DAPI), γH2AX and RAD51. The γH2AX staining marked the tracks of DSBs, and the percentage of DSB tracks that co-localized with RAD51 was determined for 100 DSB track positive cells per data point. Error bars represent the SEM of three independent experiments.
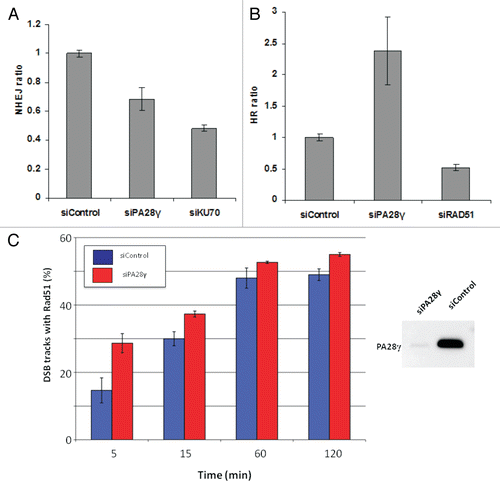
Figure 5 A fraction of PA28γ is recruited to DNA damage sites. (A) U2OS cells were treated with 400 ng/ml of NCS. At various time points following damage induction, the chromatin fraction was isolated and blotted with the indicated antibodies. (B) DNA damage was induced in U2OS cells using a focused laser microbeam. Twenty min later, the cells were treated with 0.25% NP40, fixed and co-stained with antibodies against PA28γ and γH2AX. (C) U2OS cells stably expressing GFP-PA28γ were transfected with DsRed-PCNA, and localized DNA damage was induced using a focused laser micro-beam. The cells were monitored using time-lapse imaging. Note the fraction of PA28γ that was recruited to the sites of DNA damage and co-localized with DsRed-PCNA, lasting up to 1 h following damage induction.
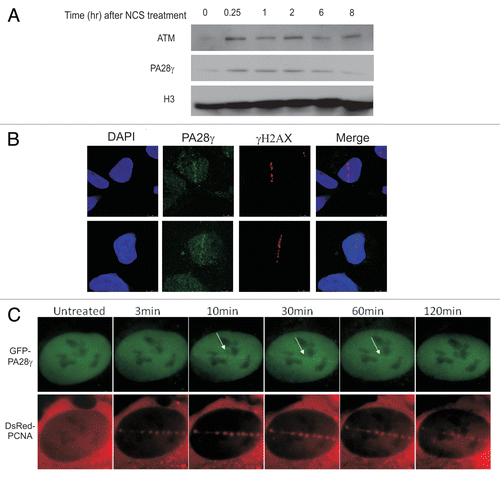
Figure 6 A fraction of PA28γ is phosphorylated by ATM in response to DNA damage. HEK293 cells were treated with NCS and cellular extracts separated on SDS-PAGE using 30 µM Phos-tag. (A) Immunoblots of PA28γ from cells treated with 400 ng/ml NCS for 30 min. The cellular extracts were or were not treated with λPPase. (B) Immunoblots of PA28γ from cells treated with various doses of NCS for 30 min. (C) Effect of the ATM inhibitor KU-55933 (ATMi) and DNA-PKs inhibitor NU-7441 (DNA-PKcs i) at 10 µM concentration on PA28γ phosphorylation, demonstrated using Phos-tag or regular gel. One hr after addition of the inhibitor, the cells were treated with 400 ng/ml of NCS for 30 min. HSC70 served as loading control, and phosphorylation of KAP-1 Citation19 represented the activation of the ATM-dependent DNA damage response. (D) Kinetics of PA28γ phosphorylation demonstrated using the Phos-tag or regular gel. The cells were treated with 400 ng/ml of NCS.
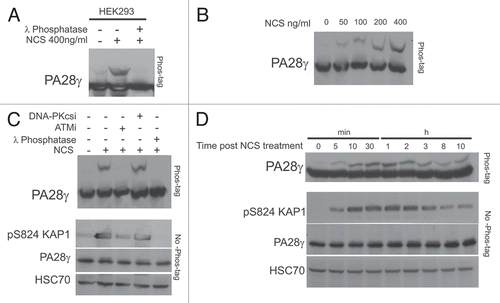
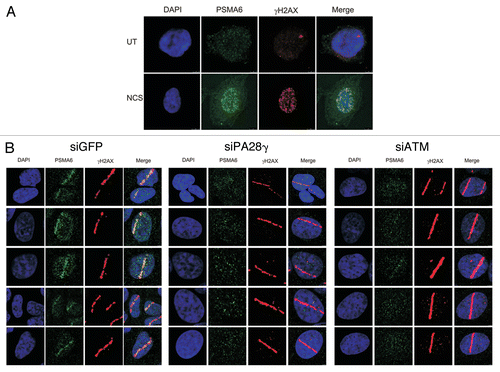
Figure 7 PA28γ- and ATM-dependent recruitment of the 20S proteasome to DNA damage sites. (A) CAL51 cells were treated with 200 ng/ml NCS for 30 min and co-stained with antibodies against the 20S core proteasome subunit PSMA6 and γH2AX. (B) U2OS cells were transfected with siRNAs against GFP or PA28γ or ATM, and localized DNA damage was induced 72 h later by a laser microbeam. Twenty min later, the cells were co-stained with antibodies against PSMA6 and γH2AX.