Figures & data
Figure 1 (A) Genomic structure of the dAven gene. The three predicted exons of dAven are represented by rectangles connected by lines representing introns. The first exon includes a 95-bp 5′-UTR (untranslated region; dark grey) and a 885-bp coding region (light grey), whereas the rest of the coding sequence is located within the second exon (grey box). The third exon is composed of a 185-bp 3′-UTR (dark grey). (B) Alignment of the predicted dAven protein sequence and Aven proteins from various vertebrate species. The predicted amino acid sequence of dAven (AY071046) was manually aligned with those of Aven proteins of human (NP_065104.1), mouse (NP_083120), bird (NP_001005791), frog (NP_001090621) and fish (NP_001038757) origins. Identical residues (white letters in black boxes) and highly conserved residues (white letters in dark grey boxes) in dAven that are conserved in at least two other species are highlighted. Other conserved residues are shown in black letters and a light grey box. (:) indicates small residues (G, A, S, P, T, D, N); (.) indicates charged residues (K, R, D, E, H); and (*) indicates polar residues (K, R, D, E, N, Q).
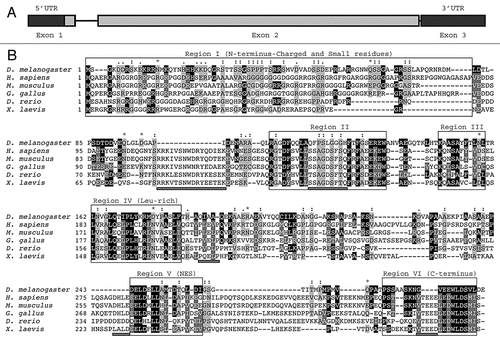
Figure 2 Expression of dAven in Drosophila S2R+ cells. (A) Immunoblot of HA-tagged dAven expressed in S2R+ cells. Expression of dAven in S2R+ cells was achieved by transfection with a pMT/V5-His plasmid coding for HA-dAven followed by CuSO4 addition to the media to induce protein production. Extracts from these cells (transfected) and mock transfected controls (mock) were analyzed by PAGE followed by immunoblotting with anti-HA antibodies. (*) indicates a non-specific band present in both samples. (B) Immunolocalization of HA-tagged dAven transiently expressed in S2R+ cells. S2R+ cells were grown in coverslips and later transfected and induced as in (A). Subsequently, cells were fixed in paraformaldehyde, permeabilized with acetone and probed with anti-HA antibodies followed by anti-Rabbit-FITC conjugates. A representative immunofluorescence image is shown. Controls with anti-HA stain on cells transfected with an empty vector were completely devoid of signal at the exposure times analyzed (data not shown). Magnification 60X. Scale bar, 5 µm.
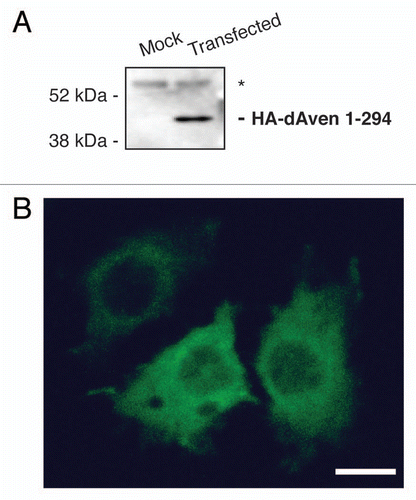
Figure 3 Embryonic expression of dAven. High resolution in situ hybridization of Drosophila embryos at various stages using an RNA-DIG labeled probe encoding an anti-sense sequence of dAven mRNA. (A) Early Drosophila embryo showing dividing nuclei (stained with DAPI and pseudo-colored in red, and indicated by arrows) and maternal dAven mRNA throughout the embryo (green). (B) Stage 5 embryo showing dAven mRNA enriched at the yolk and the yolk nuclei area (yn) and absent from the pole cells (pc) and the blastoderm nuclei (bn). (C) Embryo during gastrulation with ubiquitous distribution of dAven mRNA, including a mild enrichment at the mesoderm (m) and some zygotic transcription (asterisks). (D) Higher magnification image from the same embryo as (C) showing dAven mRNA localizing to cytoplasmic foci (white arrow) outside the nuclei (red) of cells. Zygotic transcription is seen as green dots inside the nuclei (asterisks). (E) Stage 9 embryo with dAven expression throughout the embryo, including the amnioserosa (as). (F) Stage 13 embryo showing ubiquitous dAven expression, including the ventral nerve cord (vnc), the ectoderm (ec) and the amnioserosa (as). (G) and (H) Stage 16 embryo showing dAven expression at the salivary glands (sg) and the malpighian tubules (mt); (H) is a higher magnification image of (G). In all the panels, anterior is to the right and ventral is down.
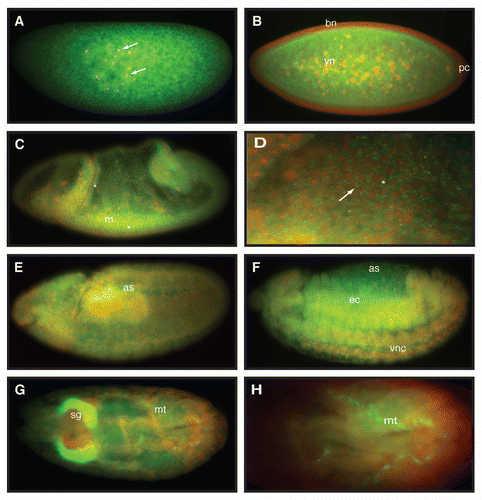
Figure 4 dAven knockdown by RNAi in Drosophila fat cells leads to smaller cells with PHH3-positive, highly condensed DNA. (A–C) UAS-AvenRNAi control (A), CG-Gal4 control (B), and CG>UAS-AvenRNAi (C) fat cells were labeled with DAPI (blue), PHH3 (green), and α-spectrin (red). PHH3 and DAPI are shown in (A′–C′) and (A″–C″), respectively. Magnified DAPI-stained fat cell nuclei from (A″–C″, dashed box) shown in (A‴–C‴). Scale bars, 10 µm.
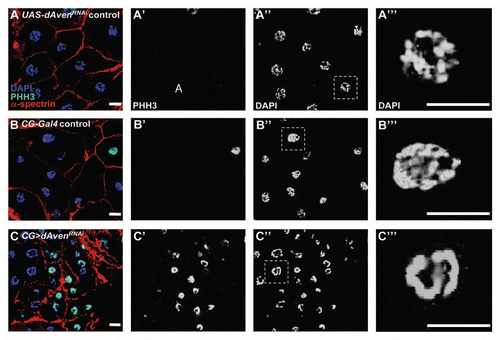
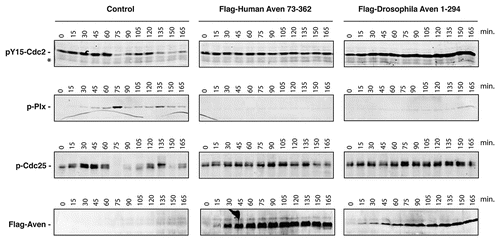
Figure 5 The dAven protein effectively blocks mitotic entry in Xenopus egg extracts. Control buffer (control) or mRNA-encoding either Flag-tagged human Aven (Flag-Human Aven 73-362) or Flag-tagged dAven (Flag-Drosophila Aven 1-294) were added to cycling Xenopus egg extracts to drive protein expression. Aliquots of extracts were then taken at the indicated times and immunoblotted with anti-phospho Tyr15-Cdc2 (pY15-Cdc2, upper panels), anti-phospho polo-like kinase-1 (pPlx, middle panels), anti-phospho Cdc25 (pCdc25, lower panels) and anti-Flag epitope (bottom panels) antibodies. (*) indicates a non-specific band detected by the anti-phospho Tyr15-Cdc2 that serves as loading control. Data shown are representative results of at least three replicates.