Figures & data
Figure 1. Presence of p53 confers resistance to cisplatin. (A) EI-H1299 cells were induced with ponasterone A to express p53 prior to treatment with increasing dosage of cisplatin. Cell viability was determined through 7-ADD staining, flow cytometry and analyzed using FlowJo software. (B) EI-H1299 cells were induced with ponasterone A to express p53 prior to treatment with 208 µM of cisplatin. Apoptosis was measured by detecting annexin V staining using flow cytometry and the data was analyzed using FlowJo software. (C) H1299 cells were induced with ponasterone A to express p53 prior to treatment with 25 µM cisplatin for 48 h. The cells were then allowed to recover in the absence of cisplatin over 12 days and were stained with crystal violet to visualize cell growth. Cytosolic extracts of the various cell lines were probed for expression of p53 (bottom panel). The area of cell growth was measured using ImageJ, and the graph (top panel) shows the change in cell growth area of the three cell lines between the induced and non-induced cells. (D) HCT-116 p53-/- cells were transfected with the various GFP-tagged p53 before 0.5 mM of cisplatin was added. Cells were counted at every 24-h period post administration of cisplatin. The percentages of transfected cells were calculated and the ratios of transfected cells in cisplatin to those in PBS were obtained. Un-normalized data used to calculate the fold change is presented in Figure S2.
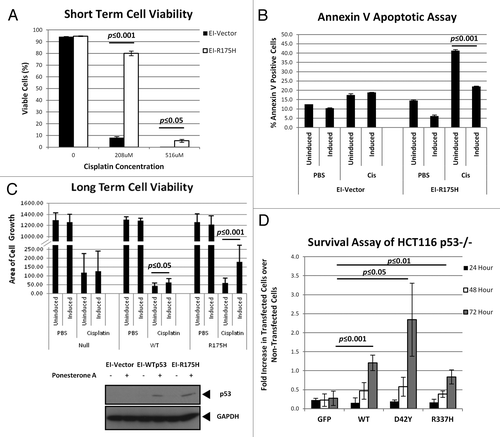
Figure 2. Different cytoplasmic levels of p53 affect cleavage of caspase-9. (A) Recombinant WT and mutant p53 were added to cytochrome-c challenged S100 lysates of HCT116 p53-/- cells and the caspase cleavage profiles were probed. (B) Endogenous p53 in HEK293 cells was knocked-down using siRNA and the S100 cytosolic extracts of the cells were induced with recombinant cytochrome-c to initiate apoptosis. (C) Immunocytochemistry showing the localization of p53R175H and p53R273H in H1299 cells stably expressing these mutants (left panel). Immunoblots of p53 and caspase-9 of the cytochrome-c challenged S100 lysates from each cell type at different time points were probed (right panel).
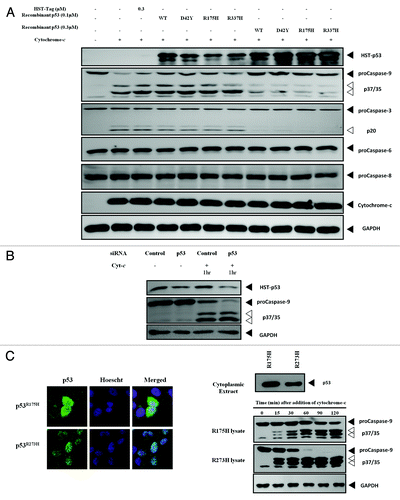
Figure 3. WT and mutant p53 interacts directly with caspase-9.(A) One µM of recombinant p53 was incubated with recombinant active caspase-9 and the caspase-9 activity was measured after 1 h incubation (left panel). Increasing concentrations of the various recombinant p53 used in (A) were incubated with recombinant active caspase-9, and the caspase-9 activity was measured after 1 h incubation (right panel). (B) Procaspase-3 was expressed using an IVT system and the recombinant active caspase-9-induced cleavage was probed in the presence of control proteins and recombinant WTp53 after 2 h incubation. (C) One µM of recombinant p53 was added to recombinant active caspase-3 (left panel) or caspase-6 (right panel) and the activities of the respective caspases were measured after 1 h incubation.
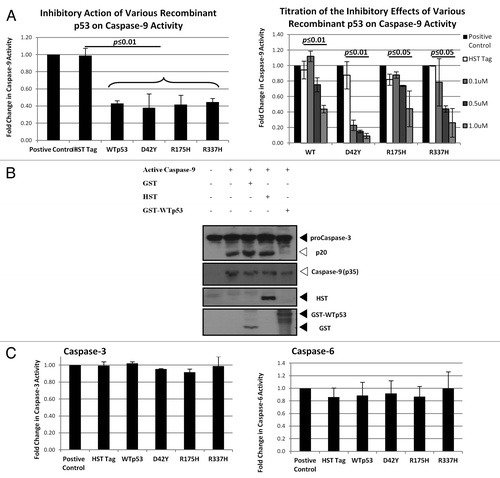
Figure 4. WTp53 interacts with the p35 fragment caspase-9.(A) p53 was expressed using an IVT system, and immunoprecipitation was performed in the presence of IVT-expressed MDM2 or recombinant active caspase-9. (B) Apoptosis was induced with dATP and rCyt-c in S100 lysates of H1299 cells stably expressing p53R175H. Antibody pairs between DO-1 (p53) and the above was added to the lysates and anti-mouse coated (donor ) and anti-rabbit (acceptor) beads were used to detect interactions between the proteins (top panel). Fluorescence signal were measured for lysates induced for apoptosis and expressed as a ratio over signal obtained for uninduced lysates for each antibody pair (bottom left panel). Immunoblots for p53 and caspase-9 was done for the lysates used for this assay (bottom right panel).
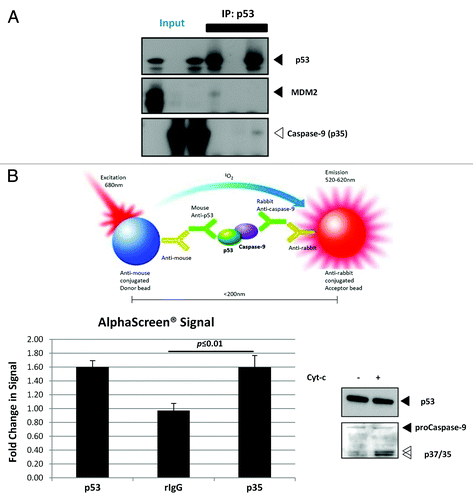
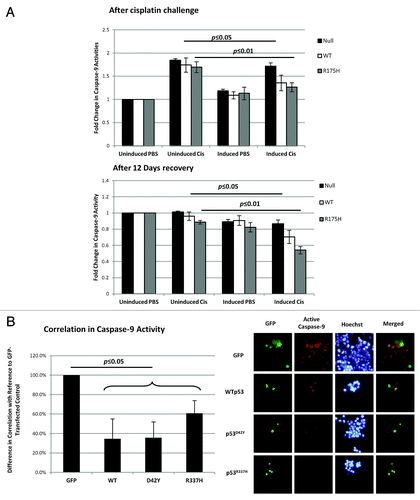
Figure 5. Resistance to cisplatin in the presence of p53 correlated with the inhibition of caspase-9 activity. (A) H1299 cells were induced with ponasterone A to express p53 prior to treatment with 25 µM cisplatin for 48 h. After 48 h with cisplatin, caspase-9 activity was measured and normalized to cell density (top panel). The cells were then allowed to recover in the absence of cisplatin over 12 d. Caspase-9 activity after the 12-d recovery period was also measured (bottom panel). (B) HCT-116 p53-/- cells were transfected with the various GFP-tagged p53 before 0.5 mM of cisplatin was added. At 72 h, substrates that are specific to the action of active caspase-9 were added to the cells to yield a fluorescent product 1 h prior to imaging. Cells positive for both GFP fluorescence and caspase-9 activity were counted and expressed as a ratio over the total number of GFP-fluorescence-positive cells (left panel). The right panel shows the representative images used to calculate the correlation percentages.