Figures & data
Table 1.SIRT1 mutation analysis of 40 human breast cancer cell lines and HeLa cell line
Table 2.SIRT1 sequence variants among 41 cancer cell lines
Figure 1. Identification of three SIRT1 mutant cancer cell lines by PCR amplification and direct sequencing. (A) Twenty-four-bp deletion of nucleotides 245–268 was found in the HeLa cell line, corresponding to an in-frame deletion of eight amino acids (R65_A72del). (B) One missense mutation 2244A > G, resulting in one amino acid change I731V, was discovered in breast cancer cell line, SUM185PE. (C) One missense mutation 2268G > T, resulting in one amino acid change D739Y, was discovered in breast cancer cell line, ZR75–30. Top row of each data represents electropherograms showing the wild-type sequence. Bottom row of each data represents electropherograms showing the mutations.
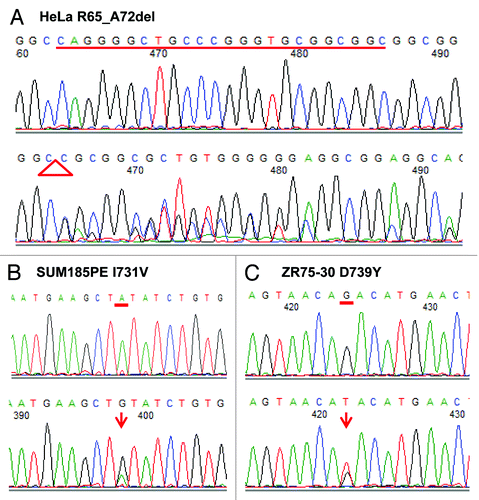
Figure 2. In vitro deacetylase activity assay of wild-type and mutant SIRT1 proteins. (A) Coomassie-stained gel showing expression and purity of wild-type and mutant SIRT1 proteins. (B) Activity of wild-type and mutant SIRT1 proteins determined using the BIOMOL assay (n = 2; mean + s.d. shown). (C) Activity of wild-type and mutant SIRT1 proteins determined using the PNC1-OPT assay with a natural amino acid substrate (n = 3; mean + s.d. shown).
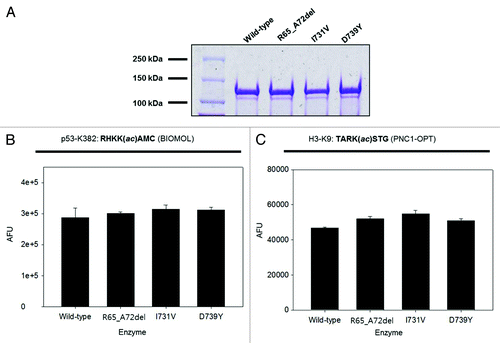
Figure 3. Telomerase activity in 293T cells overexpressing SIRT1 mutants. Cells were transfected with plasmids coding for indicated mutants or empty vector, and TRAP assays were conducted after 48 h of transfections. The graph shows mean ± s.d. from triplicate experiments.
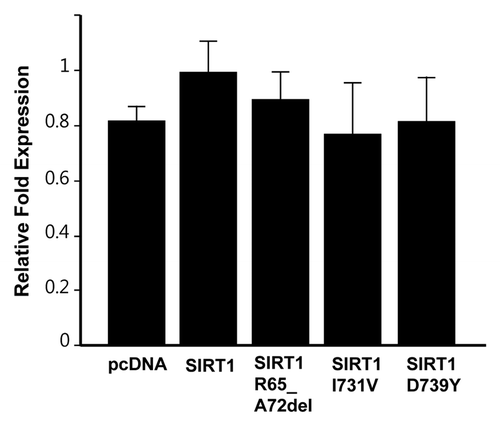
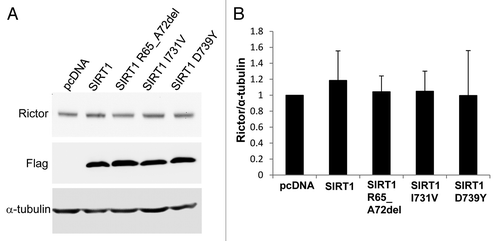
Figure 4. Western blot analysis of Rictor levels. (A) HEK293 cells were transfected with either pcDNA3.1 vector, Flag-tagged wild-type SIRT1 or Flag-tagged mutant SIRT1 constructs. Forty-eight hours post-transfection, the cells were collected and the proteins were analyzed by western blot with antibodies against Rictor, Flag and α-tubulin. (B) Bar graph showing the quantification of Rictor compared with α-tubulin. Values are means ± SE for three different sets of western blots.