Figures & data
Table 1. Effects of PN removal on the development competence of reconstructed zygotes
Figure 1. Effects of PN removal on the quality of reconstructed eight-cell embryos and the representative image of embryos in 3PN, r2PN and r1PN groups. (A) The quality of embryos at the eight-cell stage tended to decrease with the increasing number of removed PN. The ratio of high-quality embryos was significantly decreased in the r1PN group. Means with “a” or “b” are significantly different (p < 0.05). (B) No significant morphological differences were observed among the three groups before blastocyst formation; however, no high-quality blastocysts were formed in the r1PN group.
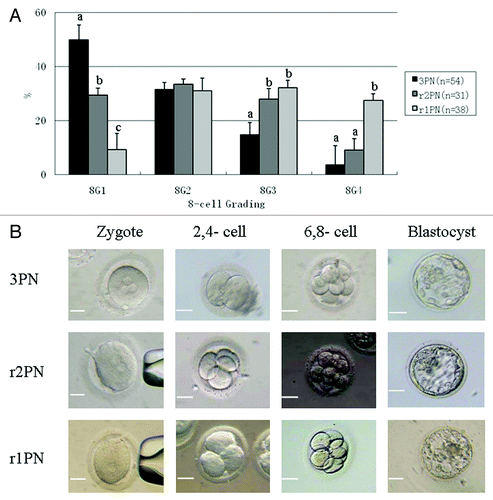
Table 2. Summary of quality scoring and ES derivation in 3PN-, r2PN- and r1PN-blastocysts
Figure 2. Representative tripolar and bipolar spindle assembly among the 3PN, r2PN and r1PN groups. (A) Tripolar spindle morphology among the three groups; (B) bipolar spindle morphology in both the r2PN and r1PN groups; (C) all of the tested zygotes in the 3PN group had a tripolar spindle assembly, whereas 88.9% (n = 18) of the tested zygotes in the r2PN group and 93.3% (n = 15) of the tested zygotes in the r1PN group had a tripolar spindle assembly.
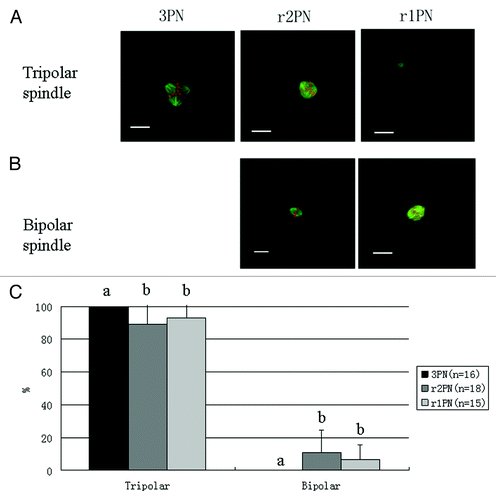
Figure 3. Analysis of chromosome ploidy using fluorescence in situ hybridization. (A) Triploid in 3PN zygote; (B) diploid in r2PN zygote; (C) haploid in r1PN zygote; (D) the loss of chromosome 18 was found in the r2PN and r1PN groups.
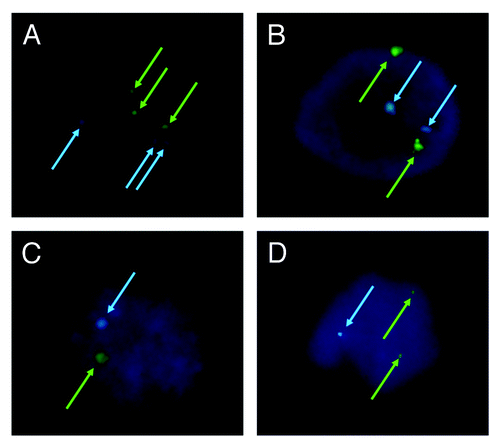
Table 3. Effects of blastomere biopsy on the blastocyst formation among three groups
Figure 4. General characteristics of hES cells from 3PN and r2PN blastocysts. (A) The inner cell mass from the blastocyst could attach and form the primary colony, which then propagated by the mechanical method in both the 3PN and r2PN groups. The distinct cell colony boundary, high nuclear/cytoplasm ratio and tightly packed colonies could be observed, and these colonies showed positive AP activity. The ES cells positively expressed a pluripotency marker (OCT4) and ES cell surface markers (SSEA3 and TRA-1–60). (B) Triploid karyotyping (69, XXY and 69, XXX) was observed in 3PN-hES cells, and diploid karyotyping (46, XY) was observed in r2PN-hES cells.
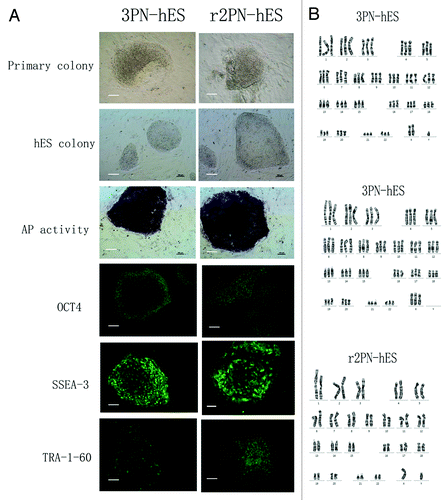
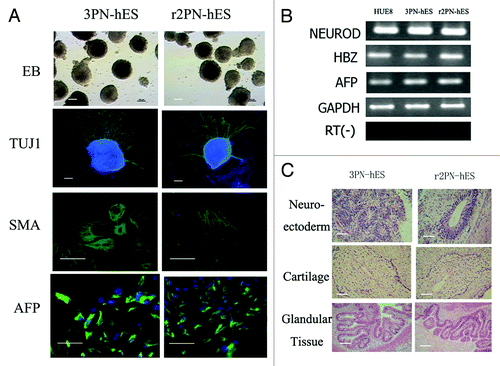
Figure 5. Differentiation ability in hES cells from the 3PN and r2PN groups. (A) EBs were formed in both types of ES cells, and TUJ-1 (ectoderm), SMA (mesoderm) and AFP (endoderm) were positively expressed in the differentiated EBs. (B) Genes from the endoderm (AFP), mesoderm (HBZ) and ectoderm (NEUROD) were identified in differentiated EB clumps. (C) Tissues from the three embryonic germ layers were identified in teratomas: neuroectoderm (ectoderm), cartilage (mesoderm), glandular tissue (endoderm).