Figures & data
Figure 1. Overall structure of RNF8N-111. (A) Ribbon representation of the structure of RNF168N-111. The enlarged view of the interaction of helix α1 with the followed loop is shown on the right side. Amino acid residues are shown in a stick mode. Zinc ions are shown in a sphere mode. The core RING domain, the fragments preceding and following the RING domain are colored green, yellow and cyan, respectively. (B) Structure-based sequence alignment of RNF168 RING domain, TRAF6 RING domain, CHIP U-box domain and RNF8 RING domain. The conserved residues are in red and strictly conserved residues are highlighted red. The residues involved in the interaction with UBC13 on the central α helix are labeled by diamond. M20 and S48 of RNF168 and corresponding residues of TRAF6, CHIP and RNF8 are labeled by circle and triangle, respectively.
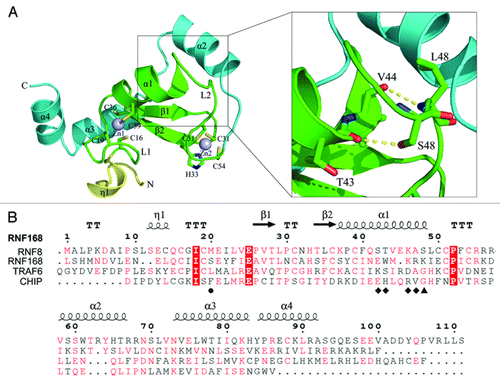
Figure 2. Comparison of the structure of RNF168N-111 with TRAF6, CHIP and RNF8. Superposition the structure of RNF168N-111 to (A) TRAF6 (PDB code: 3HCT), (B) CHIP (PDB code: 2C2V), (C) RNF8 (PDB code: 4EPO). Residues are shown in a stick mode. Metal ions are shown in a sphere mode. RNF168, TRAF6, CHIP and RNF8 are colored green, cyan, pink and brown, respectively.
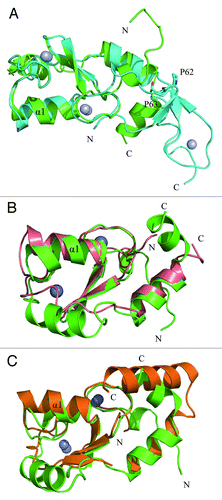
Figure 3. Structural basis for the lack of interaction between RNF168N-111 and UBC13. (A) Comparison of the binding of loop L1 of TRAF6 (left panel), CHIP (middle panel) and RNF8 (right panel) to the hydrophobic biding pocket on UBC13 surface with RNF168N-111. (B) Comparison of the binding of the C-terminal end of the central α helix of TRAF6 (left panel), CHIP (middle panel) and RNF8 (right panel) to the hydrophobic pocket on UBC13 surface with RNF168N-111. (C) Comparison of the interaction between UBC13 and TRAF6 (left panel), CHIP (middle panel) and RNF8 (right panel) to RNF168N-111. RNF168, TRAF6, CHIP and RNF8 are shown in a ribbon mode and colored green, cyan, pink and brown, respectively. Residues are shown in a stick mode. UBC13 is shown in surface representation in (A and B). White, blue and red regions indicate neutral areas, positively charged areas and negatively charged areas, respectively. UBC13 is shown in a ribbon mode and colored yellow in (C). The PDB codes of TRAF6, CHIP and RNF8 in complex with UBC13 are 3HCT, 2C2V and 4EPO, respectively.
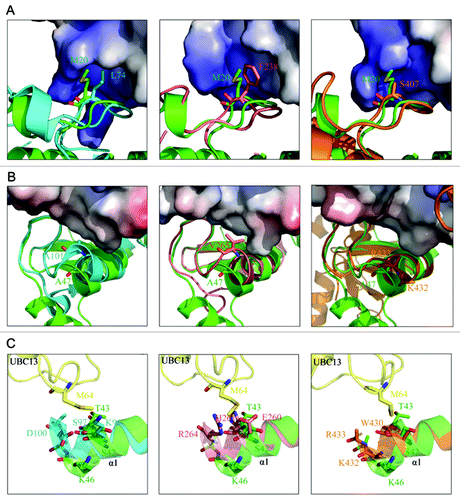
Figure 4. RNF168 does not stably associate with UBC13. (A) The interaction between RNF168N-111 and UBC13 analyzed by GST pull-down assay at 4°C and 37°C. (B) Agarose conjugated with bacterially expressed and purified GST-UBC13 or MBP-UBC13 proteins, and their corresponding M64A mutants, were incubated with lysates derived from cells expressing Flag-RNF8 or Flag-RNF168 for 4 h at 4°C with gentle agitation. Thereafter, agarose beads were washed three times with NETN buffer and proteins were separated by SDS-PAGE. Western blotting experiments were done using indicated antibodies. (C) MBP-RNF8 or MBP-RNF168 protein were incubated with lysates derived from cells expressing myc-tagged UBC13 wild-type (WT) or its M64A mutant. Bound complexes were washed and analyzed using anti-myc (9E10) antibodies. (D) GST-UBC13 proteins were incubated with lysates derived from irradiated cells or untreated cells. Bound proteins were analyzed using indicated antibodies. (E) 293T cells were co-transfected with indicated constructs. Cells were subsequently lysed using NETN buffer, and streptavidin beads were used to precipitate UBC13 protein complexes. Proteins were separated by SDS-PAGE and were analyzed by western blotting using indicated antibodies.
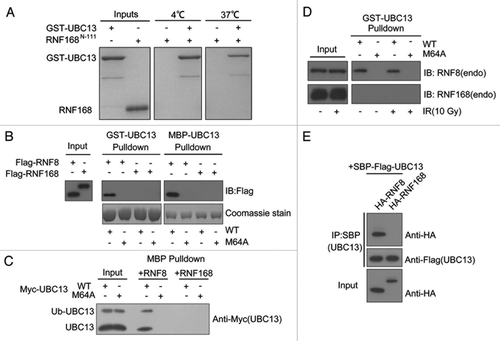
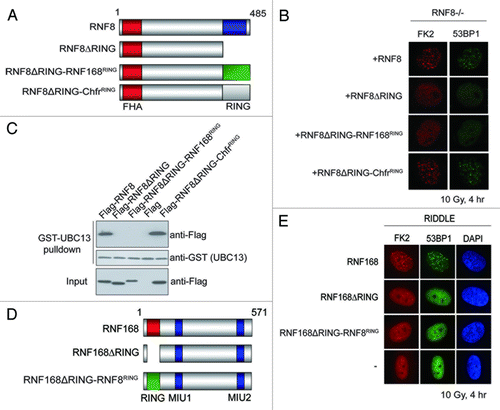
Figure 5. RNF8 and RNF168 RING domains are not functionally interchangeable. (A and D) Schematic illustrations of RNF8 and RNF168 chimeric constructs. (B and E) RNF8−/− MEFs or RIDDLE cells (RNF168 mutant cells) reconstituted with indicated constructs were irradiated (10 Gy) and were processed for immunofluorescent experiments to assess FK2 and 53BP1 foci formation. (C) GST-UBC13 interacted with RNF8 and Chfr RING but not RNF168 RING. Agarose beads conjugated with GST-UBC13 proteins were incubated with lysates derived from cells expressing indicated proteins. Bound protein complexes were separated by SDS-PAGE and analyzed by western blotting using indicated antibodies.