Figures & data
Figure 1. The C-terminal region of Chk1 contains a predicted kinase-associated 1 (KA1) domain with a unique C-terminal extension (CTE). (A) DISOPRED generated disorder plot of S. pombe Chk1. The horizontal line at 5% represents the order/disorder threshold. (B) PSIPRED secondary structure prediction of the KA1 domain (βαββββα) for S. pombe Chk1, (C) human Chk1, (D) Mouse MARK3 kinase. Note that Chk1 has an extension containing β6. α-helices (H) are green cylinders, β-sheets (E) are yellow arrows and black lines are coiled regions (C). Cyan bars represent the confidence of the prediction (0–100%). The Chk1 residues boxed by the red line are the most conserved residues that when mutated can either activate or inactivate Chk1 homologs.
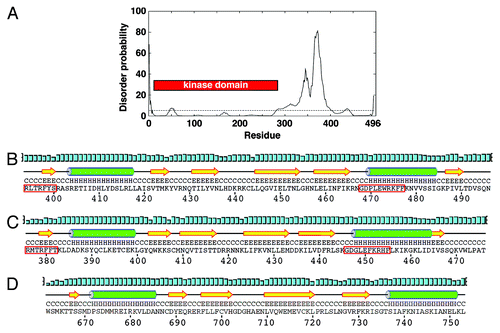
Figure 2. Mutations within the C-terminal regulatory domain ablate Chk1 function. (A) Clonogenic survival to UV-C irradiation for the indicted strains. (B) Ten-fold serial dilutions of the indicated strains were spotted onto YES agar containing no drug and 0.01% MMS and grown at 30°C for 4 d. The MMS and UV-C sensitive strains are similarly sensitive to all DNA damaging agents tested. (C) Non-functional chk1 alleles display the “cut” phenotype (arrowed) after treatment with DNA damaging agents, which is characteristic of checkpoint failure; chk1Δ, Δ½KA1 and IV488–489AA mutants are shown as examples. Under the same conditions, wild-type cells elongate due to the Chk1-dependent cell cycle arrest. Agents used are UV-C (150 J/m2), 2 h after irradiation and a 4 h incubation in the following drugs: 0.01% MMS, 0.5 mU Bleomycin (Bleo), 10 µM Camptothecin (CPT) and 1mM mechlorethamine hydrochloride (HN2).
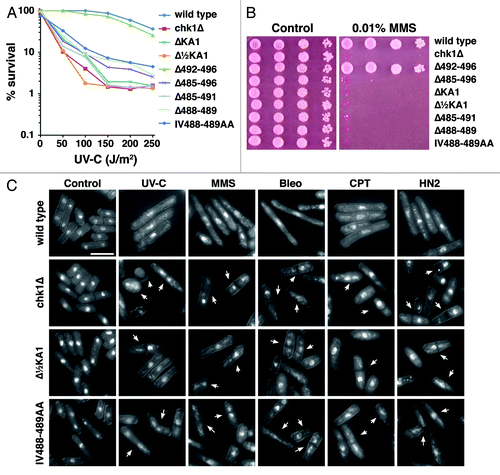
Figure 3. Non-functional Chk1 mutants fail to undergo activating phosphorylation. (A and B) Western blotting (anti-HA for Chk1, anti-tubulin as a loading control) for the indicated strains either mock irradiated, or irradiated with 150 J/m2 UV-C. Activating phosphorylation is seen as a retarded mobility on SDS-PAGE. Note that the ΔKA1 and Δ488–489 mutants show no detectable protein expression, but (C) show normal mRNA levels by northern blotting (28S rRNA is shown as a loading control). (D) The indicated Chk1 mutants were expressed in cells lacking the Chk1 S345 phosphatase Dis2, and either mock-treated or incubated with 0.01% MMS for 7 h. The non-functional alleles still fail to show activating phosphorylation.
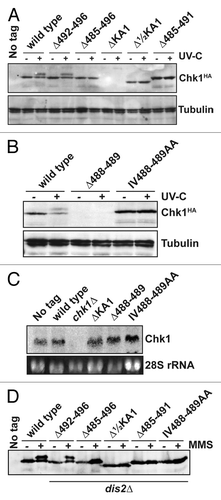
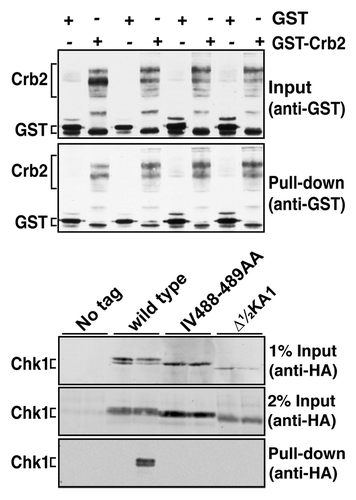
Figure 4. Non-functional Chk1 mutants fail to interact with Crb2. Strains expressing the indicated Chk1 mutants were transformed with plasmids expressing GST or GST-Crb2 from the nmt1 promoter. Expression from nmt1 was induced by thiamine withdrawal for 12 h at 30°C, whereupon cells were then incubated with 0.01% MMS for a further 7 h. The top panels show the input (50 µg total extract) and recovered (on glutathione magnetic beads) GST and GST-Crb2, as well as the endogenous S. pombe GST that runs slightly faster than the exogenous GST. Lower panels show 1% or 2% of the input for Chk1 (50 µg or 100 µg of total extract) and the Chk1 (wild-type only) recovered on glutathione magnetic beads.